The graph information of MGIVmi1 components from NZ_ACYV01000005 |
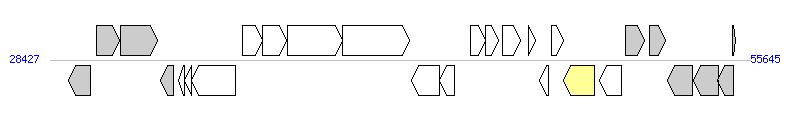 |
| Complete gene list of MGIVmi1 from NZ_ACYV01000005 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | VMD_RS14830 | 29140..29988 [-], 849 | MurR/RpiR family transcriptional regulator | |
| 2 | murQ | 30219..31121 [+], 903 | N-acetylmuramic acid 6-phosphate etherase | |
| 3 | VMD_RS14820 | 31152..32597 [+], 1446 | PTS N-acetylmuramic acid transporter subunits IIBC | |
| 4 | VMD_RS14815 | 32739..33227 [-], 489 | TIGR00645 family protein | |
| 5 | VMD_RS14810 | 33427..33651 [-], 225 | hypothetical protein | |
| 6 | VMD_RS14805 | 33652..33936 [-], 285 | DNA-binding protein | |
| 7 | VMD_RS14800 | 33933..35627 [-], 1695 | DUF927 domain-containing protein | |
| 8 | VMD_RS14795 | 35896..36687 [+], 792 | hypothetical protein | |
| 9 | VMD_RS14790 | 36680..37618 [+], 939 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | |
| 10 | VMD_RS14785 | 37657..39798 [+], 2142 | DNA methyltransferase | |
| 11 | VMD_RS14780 | 39801..42401 [+], 2601 | hypothetical protein | |
| 12 | VMD_RS14775 | 42475..43563 [-], 1089 | ImmA/IrrE family metallo-endopeptidase | |
| 13 | VMD_RS14770 | 43569..44165 [-], 597 | hypothetical protein | |
| 14 | VMD_RS14765 | 44765..45352 [+], 588 | hypothetical protein | |
| 15 | VMD_RS0118280 | 45349..45879 [+], 531 | hypothetical protein | |
| 16 | VMD_RS14760 | 46008..46721 [+], 714 | hypothetical protein | |
| 17 | VMD_RS0118285 | 47018..47302 [+], 285 | hypothetical protein | |
| 18 | VMD_RS0118290 | 47468..47797 [-], 330 | XRE family transcriptional regulator | |
| 19 | VMD_RS0118295 | 47909..48397 [+], 489 | hypothetical protein | |
| 20 | VMD_RS14755 | 48389..49594 [-], 1206 | site-specific integrase | Integrase |
| 21 | VMD_RS14750 | 49779..50645 [-], 867 | YicC family protein | |
| 22 | VMD_RS14745 | 50809..51525 [+], 717 | ribonuclease PH | |
| 23 | VMD_RS14740 | 51724..52365 [+], 642 | orotate phosphoribosyltransferase | |
| 24 | msbB | 52451..53428 [-], 978 | lauroyl-Kdo(2)-lipid IV(A) myristoyltransferase | |
| 25 | VMD_RS14730 | 53447..54403 [-], 957 | lipid A biosynthesis lauroyl acyltransferase | |
| 26 | VMD_RS14725 | 54400..54990 [-], 591 | nucleoid occlusion factor SlmA | |
| 27 | VMD_RS20835 | 54946..55086 [+], 141 | lipid A biosynthesis lauroyl acyltransferase | |