The graph information of ICEhptfs4b components from NC_011333 |
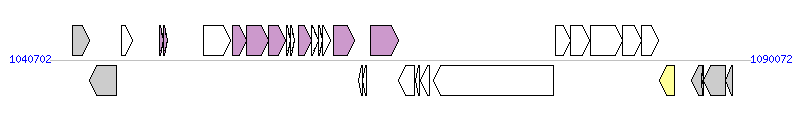 |
| Complete gene list of ICEhptfs4b from NC_011333 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | HPG27_RS04975 | 1042313..1043482 [+], 1170 | class I SAM-dependent methyltransferase | |
| 2 | HPG27_RS04980 | 1043492..1045363 [-], 1872 | mechanosensitive ion channel family protein | |
| 3 | HPG27_RS04985 | 1045764..1046528 [+], 765 | hypothetical protein | |
| 4 | HPG27_RS04995 | 1048432..1048734 [+], 303 | hypothetical protein | VirB2, T4SS component |
| 5 | HPG27_RS05000 | 1048735..1048995 [+], 261 | hypothetical protein | VirB3, T4SS component |
| 6 | HPG27_RS05010 | 1051500..1053434 [+], 1935 | type IA DNA topoisomerase | |
| 7 | HPG27_RS05015 | 1053564..1054550 [+], 987 | competence protein | VirB8, T4SS component |
| 8 | HPG27_RS05020 | 1054550..1056091 [+], 1542 | conjugal transfer protein | VirB9, T4SS component |
| 9 | HPG27_RS05025 | 1056088..1057323 [+], 1236 | competence protein | VirB10, T4SS component |
| 10 | HPG27_RS05030 | 1057393..1057689 [+], 297 | hypothetical protein | |
| 11 | HPG27_RS05035 | 1057679..1057942 [+], 264 | hypothetical protein | |
| 12 | HPG27_RS05045 | 1058237..1059178 [+], 942 | hypothetical protein | VirB11, T4SS component |
| 13 | HPG27_RS05050 | 1059175..1059630 [+], 456 | hypothetical protein | |
| 14 | HPG27_RS05055 | 1059627..1059917 [+], 291 | hypothetical protein | |
| 15 | HPG27_RS05060 | 1059918..1060475 [+], 558 | hypothetical protein | |
| 16 | HPG27_RS05065 | 1060731..1062203 [+], 1473 | type IV secretory system conjugative DNA transfer family protein | T4CP |
| 17 | HPG27_RS05070 | 1062466..1062747 [-], 282 | type II toxin-antitoxin system mRNA interferase toxin, RelE/StbE family | |
| 18 | HPG27_RS05075 | 1062740..1063054 [-], 315 | hypothetical protein | |
| 19 | HPG27_RS05080 | 1063336..1065246 [+], 1911 | hypothetical protein | Relaxase, MOBP Family |
| 20 | HPG27_RS05085 | 1065271..1066443 [-], 1173 | hypothetical protein | |
| 21 | HPG27_RS05090 | 1066475..1066759 [-], 285 | hypothetical protein | |
| 22 | HPG27_RS05095 | 1066842..1067498 [-], 657 | ParA family protein | |
| 23 | HPG27_RS05100 | 1067764..1076190 [-], 8427 | helicase | |
| 24 | HPG27_RS05105 | 1076345..1077382 [+], 1038 | hypothetical protein | |
| 25 | HPG27_RS05110 | 1077383..1078777 [+], 1395 | hypothetical protein | |
| 26 | HPG27_RS05115 | 1078838..1081051 [+], 2214 | hypothetical protein | |
| 27 | HPG27_RS05120 | 1081055..1082440 [+], 1386 | hypothetical protein | |
| 28 | HPG27_RS05125 | 1082440..1083642 [+], 1203 | hypothetical protein | |
| 29 | HPG27_RS05130 | 1083705..1084781 [-], 1077 | site-specific integrase | Integrase |
| 30 | HPG27_RS05160 | 1085950..1086699 [-], 750 | neuraminyllactose-binding hemagglutinin HpaA | VF |
| 31 | HPG27_RS08055 | 1086686..1086826 [-], 141 | hypothetical protein | |
| 32 | HPG27_RS05165 | 1086783..1088309 [-], 1527 | glutamine-hydrolyzing GMP synthase | |
| 33 | HPG27_RS05170 | 1088320..1088808 [-], 489 | hypothetical protein | |
