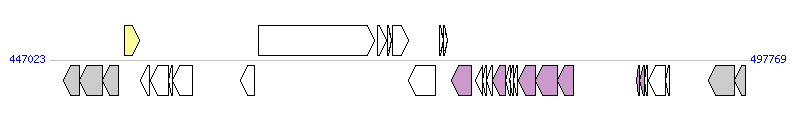The graph information of ICEhptfs4a components from NC_011498 |
 |
| Complete gene list of ICEhptfs4a from NC_011498 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | HPP12_RS02245 | 448002..449183 [-], 1182 | restriction endonuclease subunit S | |
| 2 | HPP12_RS02250 | 449176..450807 [-], 1632 | SAM-dependent DNA methyltransferase | |
| 3 | HPP12_RS02255 | 450800..451999 [-], 1200 | type I restriction endonuclease subunit R | |
| 4 | HPP12_RS02260 | 452423..453496 [+], 1074 | integrase | Integrase |
| 5 | HPP12_RS02265 | 453578..454261 [-], 684 | hypothetical protein | |
| 6 | HPP12_RS02270 | 454334..455596 [-], 1263 | P-type conjugative transfer protein TrbL | |
| 7 | HPP12_RS02275 | 455598..455876 [-], 279 | hypothetical protein | |
| 8 | HPP12_RS02280 | 455928..457328 [-], 1401 | hypothetical protein | |
| 9 | HPP12_RS02295 | 460810..461832 [-], 1023 | DUF1738 domain-containing protein | |
| 10 | HPP12_RS02300 | 462135..470522 [+], 8388 | DEAD/DEAH box helicase | |
| 11 | HPP12_RS02305 | 470785..471441 [+], 657 | ParA family protein | |
| 12 | HPP12_RS02310 | 471523..471807 [+], 285 | hypothetical protein | |
| 13 | HPP12_RS02315 | 471839..473017 [+], 1179 | hypothetical protein | |
| 14 | HPP12_RS02320 | 473042..474955 [-], 1914 | hypothetical protein | |
| 15 | HPP12_RS02325 | 475253..475567 [+], 315 | hypothetical protein | |
| 16 | HPP12_RS02330 | 475560..475841 [+], 282 | type II toxin-antitoxin system mRNA interferase toxin, RelE/StbE family | |
| 17 | HPP12_RS02335 | 476105..477577 [-], 1473 | type IV secretory system conjugative DNA transfer family protein | T4CP |
| 18 | HPP12_RS02340 | 477879..478391 [-], 513 | hypothetical protein | |
| 19 | HPP12_RS02345 | 478392..478682 [-], 291 | hypothetical protein | |
| 20 | HPP12_RS02350 | 478679..479134 [-], 456 | hypothetical protein | |
| 21 | HPP12_RS02355 | 479131..480072 [-], 942 | hypothetical protein | VirB11, T4SS component |
| 22 | HPP12_RS02360 | 480072..480371 [-], 300 | hypothetical protein | |
| 23 | HPP12_RS02365 | 480368..480631 [-], 264 | hypothetical protein | |
| 24 | HPP12_RS02370 | 480624..480917 [-], 294 | hypothetical protein | |
| 25 | HPP12_RS02375 | 480987..482252 [-], 1266 | hypothetical protein | VirB10, T4SS component |
| 26 | HPP12_RS02380 | 482252..483784 [-], 1533 | conjugal transfer protein | VirB9, T4SS component |
| 27 | HPP12_RS02385 | 483784..484953 [-], 1170 | hypothetical protein | VirB8, T4SS component |
| 28 | HPP12_RS02405 | 489522..489788 [-], 267 | hypothetical protein | VirB3, T4SS component |
| 29 | HPP12_RS02410 | 489789..490091 [-], 303 | hypothetical protein | VirB2, T4SS component |
| 30 | HPP12_RS08340 | 490088..490372 [-], 285 | hypothetical protein | |
| 31 | HPP12_RS02415 | 490433..491611 [-], 1179 | hypothetical protein | |
| 32 | HPP12_RS02420 | 491618..491911 [-], 294 | hypothetical protein | |
| 33 | HPP12_RS02435 | 494743..496638 [-], 1896 | DUF115 domain-containing protein | |
| 34 | HPP12_RS02440 | 496664..497431 [-], 768 | hypothetical protein | |