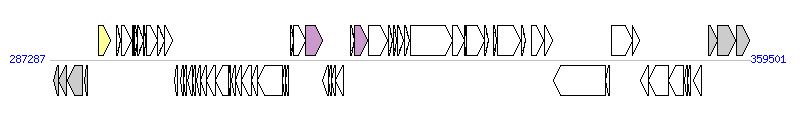The graph information of ICEEa1 components from CP014805 |
 |
| Incomplete gene list of ICEEa1 from CP014805 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | A2T74_01250 | 287689..288162 [-], 474 | hypothetical protein | |
| 2 | A2T74_01255 | 288167..288967 [-], 801 | endonuclease | |
| 3 | A2T74_01260 | 289057..290613 [-], 1557 | ribonuclease G | |
| 4 | A2T74_01265 | 290836..291132 [-], 297 | integration host factor subunit beta | |
| 5 | A2T74_00005 | 3930529..3059 [-], -3927469 | hypothetical protein | |
| 6 | A2T74_01275 | 292287..293537 [+], 1251 | integrase | Integrase |
| 7 | A2T74_01280 | 294189..294581 [+], 393 | single-stranded DNA-binding protein | |
| 8 | A2T74_01285 | 294627..295700 [+], 1074 | alpha/beta hydrolase | |
| 9 | A2T74_01290 | 295753..295947 [+], 195 | DNA-binding protein | |
| 10 | A2T74_01295 | 295974..296312 [+], 339 | molybdenum ABC transporter ATP-binding protein | |
| 11 | A2T74_01300 | 296337..296945 [+], 609 | prtrc system protein e | |
| 12 | A2T74_01305 | 296963..297181 [+], 219 | PRTRC system protein C | |
| 13 | A2T74_01310 | 297200..298363 [+], 1164 | hypothetical protein | |
| 14 | A2T74_01315 | 298360..299082 [+], 723 | PRTRC system protein B | |
| 15 | A2T74_01320 | 299079..299885 [+], 807 | hypothetical protein | |
| 16 | A2T74_01325 | 300101..300487 [-], 387 | histidine kinase | |
| 17 | A2T74_01330 | 300770..301063 [-], 294 | DNA-binding protein | |
| 18 | A2T74_01335 | 301044..301355 [-], 312 | DNA-binding protein | |
| 19 | A2T74_01340 | 301497..301940 [-], 444 | hypothetical protein | |
| 20 | A2T74_01345 | 301975..302256 [-], 282 | molybdenum ABC transporter permease | |
| 21 | A2T74_01350 | 302371..302820 [-], 450 | conjugal transfer protein TraQ | |
| 22 | A2T74_01355 | 302832..303392 [-], 561 | conjugal transfer protein TraO | |
| 23 | A2T74_01360 | 303414..304316 [-], 903 | conjugal transfer protein TrbN | |
| 24 | A2T74_01365 | 304342..305682 [-], 1341 | conjugal transfer protein TraM | |
| 25 | A2T74_01370 | 305669..305956 [-], 288 | nitrogen regulatory IIA protein | |
| 26 | A2T74_01375 | 305977..306408 [-], 432 | hypothetical protein | |
| 27 | A2T74_01380 | 306420..307043 [-], 624 | conjugal transfer protein | |
| 28 | A2T74_01385 | 307068..308063 [-], 996 | conjugal transfer protein | |
| 29 | A2T74_01390 | 308066..308698 [-], 633 | hypothetical protein | |
| 30 | A2T74_01395 | 308726..311227 [-], 2502 | conjugal transfer protein TraG | |
| 31 | A2T74_01400 | 311224..311556 [-], 333 | conjugal transfer protein TraF | |
| 32 | A2T74_01405 | 311569..311874 [-], 306 | conjugal transfer protein | |
| 33 | A2T74_01410 | 312079..312363 [+], 285 | transcriptional regulator | |
| 34 | A2T74_01415 | 312421..313626 [+], 1206 | hypothetical protein | |
| 35 | A2T74_01420 | 313629..315383 [+], 1755 | hypothetical protein | T4CP |
| 36 | A2T74_01425 | 315421..316053 [-], 633 | conjugal transfer protein TraD | |
| 37 | A2T74_01430 | 316056..316328 [-], 273 | hypothetical protein | |
| 38 | A2T74_01435 | 316340..316777 [-], 438 | conjugal transfer protein TraB | |
| 39 | A2T74_01440 | 316783..317550 [-], 768 | conjugal transfer protein TraA | |
| 40 | A2T74_01445 | 318290..318733 [+], 444 | hypothetical protein | |
| 41 | A2T74_01450 | 318730..320019 [+], 1290 | relaxase | Relaxase, MOBP Family |
| 42 | A2T74_01455 | 320121..322121 [+], 2001 | conjugal transfer protein TraG | |
| 43 | A2T74_01460 | 322233..322643 [+], 411 | hypothetical protein | |
| 44 | A2T74_01465 | 322735..322998 [+], 264 | hypothetical protein | |
| 45 | A2T74_01470 | 323118..323801 [+], 684 | hypothetical protein | |
| 46 | A2T74_01475 | 323813..324421 [+], 609 | hypothetical protein | |
| 47 | A2T74_01480 | 324443..328837 [+], 4395 | acriflavine resistance protein B | |
| 48 | A2T74_01485 | 328858..330054 [+], 1197 | efflux transporter periplasmic adaptor subunit | |
| 49 | A2T74_01490 | 330035..330259 [+], 225 | hypothetical protein | |
| 50 | A2T74_01495 | 330270..332108 [+], 1839 | exopolyphosphatase | |
| 51 | A2T74_01500 | 332139..332561 [+], 423 | hypothetical protein | |
| 52 | A2T74_01505 | 333018..333383 [+], 366 | hypothetical protein | |
| 53 | A2T74_01510 | 333456..335861 [+], 2406 | energy transducer TonB | |
| 54 | A2T74_01515 | 335924..336364 [+], 441 | hypothetical protein | |
| 55 | A2T74_01520 | 336985..338211 [+], 1227 | MFS transporter | |
| 56 | A2T74_01525 | 338261..339085 [+], 825 | tetracycline regulation of excision, RteC | |
| 57 | A2T74_01530 | 339193..344622 [-], 5430 | DNA methylase | |
| 58 | A2T74_01535 | 344609..345046 [-], 438 | hypothetical protein | |
| 59 | A2T74_01540 | 345233..347362 [+], 2130 | hypothetical protein | |
| 60 | A2T74_01545 | 347381..348139 [+], 759 | hypothetical protein | |
| 61 | A2T74_01550 | 348169..349050 [-], 882 | DNA-binding protein | |
| 62 | A2T74_01555 | 349040..351133 [-], 2094 | DNA topoisomerase III | |
| 63 | A2T74_01560 | 351136..352617 [-], 1482 | hypothetical protein | |
| 64 | A2T74_01565 | 352717..353010 [-], 294 | excisionase | |
| 65 | A2T74_01570 | 353039..353389 [-], 351 | DNA-binding protein | |
| 66 | A2T74_01575 | 353656..354501 [-], 846 | tetracycline regulation of excision, RteC | |
| 67 | A2T74_01580 | 355208..356128 [+], 921 | sugar kinase | |
| 68 | A2T74_01585 | 356171..358012 [+], 1842 | peptidylprolyl isomerase | |
| 69 | A2T74_01590 | 358078..359451 [+], 1374 | peptidylprolyl isomerase | |