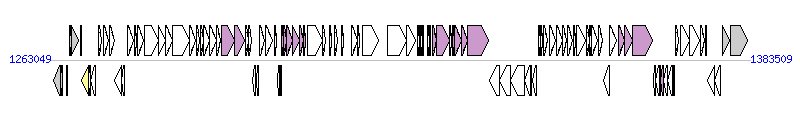The graph information of ICEAmaAS2 components from NC_021714 |
 |
| Incomplete gene list of ICEAmaAS2 from NC_021714 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | I636_RS05655 | 1263705..1264778 [-], 1074 | HlyD family secretion protein | |
| 2 | I636_RS05660 | 1264775..1265230 [-], 456 | MarR family transcriptional regulator | |
| 3 | I636_RS05665 | 1265929..1266117 [-], 189 | hypothetical protein | |
| 4 | I636_RS05670 | 1266468..1266788 [+], 321 | XRE family transcriptional regulator | |
| 5 | I636_RS05675 | 1266781..1268010 [+], 1230 | type II toxin-antitoxin system HipA family toxin | |
| 6 | I636_RS05680 | 1268227..1268421 [+], 195 | DNA-binding protein | |
| 7 | I636_RS05685 | 1268433..1269674 [-], 1242 | DUF4102 domain-containing protein | Integrase |
| 8 | I636_RS05690 | 1269676..1269945 [-], 270 | hypothetical protein | |
| 9 | I636_RS05695 | 1269948..1270922 [-], 975 | rod shape determination protein | |
| 10 | I636_RS05700 | 1271387..1271833 [+], 447 | hypothetical protein | |
| 11 | I636_RS05705 | 1272198..1273166 [+], 969 | calcium:sodium antiporter | |
| 12 | I636_RS05710 | 1273235..1274068 [+], 834 | mechanosensitive ion channel family protein | |
| 13 | I636_RS05715 | 1274077..1275345 [-], 1269 | translesion error-prone DNA polymerase V subunit UmuC | |
| 14 | I636_RS05720 | 1275353..1275802 [-], 450 | translesion error-prone DNA polymerase V autoproteolytic subunit | |
| 15 | I636_RS05725 | 1276442..1277347 [+], 906 | DNA polymerase III subunit epsilon | |
| 16 | I636_RS05730 | 1277561..1277860 [+], 300 | hypothetical protein | |
| 17 | I636_RS05735 | 1278178..1279146 [+], 969 | WYL domain-containing protein | |
| 18 | I636_RS05740 | 1279260..1281689 [+], 2430 | SAM-dependent DNA methyltransferase | |
| 19 | I636_RS05745 | 1281689..1282957 [+], 1269 | restriction endonuclease subunit S | |
| 20 | I636_RS05750 | 1282967..1284130 [+], 1164 | DUF4268 domain-containing protein | |
| 21 | I636_RS05755 | 1284145..1287045 [+], 2901 | type I restriction endonuclease subunit R | |
| 22 | I636_RS05760 | 1287068..1288153 [+], 1086 | ATP-binding region, ATPase-like protein | |
| 23 | I636_RS05765 | 1288176..1288727 [+], 552 | MarR family transcriptional regulator | |
| 24 | I636_RS05770 | 1289032..1289400 [+], 369 | metal-dependent hydrolase | |
| 25 | I636_RS05775 | 1289410..1290564 [+], 1155 | SAVED domain-containing protein | |
| 26 | I636_RS05780 | 1290557..1291747 [+], 1191 | nucleotidyltransferase | |
| 27 | I636_RS05785 | 1291748..1292461 [+], 714 | hypothetical protein | |
| 28 | I636_RS05790 | 1292578..1294728 [+], 2151 | metal-dependent phosphohydrolase | Relaxase, MOBH Family |
| 29 | I636_RS05795 | 1294777..1296600 [+], 1824 | hypothetical protein | TraD_F, T4SS component |
| 30 | I636_RS05800 | 1296610..1297170 [+], 561 | hypothetical protein | |
| 31 | I636_RS05805 | 1297157..1297792 [+], 636 | DUF4400 domain-containing protein | |
| 32 | I636_RS05810 | 1297819..1298406 [-], 588 | hypothetical protein | |
| 33 | cadR | 1298568..1298975 [-], 408 | Cd(II)/Pb(II)-responsive transcriptional regulator | |
| 34 | I636_RS05820 | 1299071..1299967 [+], 897 | cation transporter | |
| 35 | I636_RS05825 | 1300133..1301296 [+], 1164 | hypothetical protein | |
| 36 | I636_RS05835 | 1301756..1302082 [+], 327 | sel1 repeat family protein | |
| 37 | I636_RS05840 | 1302136..1302468 [-], 333 | hypothetical protein | |
| 38 | I636_RS05845 | 1302677..1302889 [-], 213 | hypothetical protein | |
| 39 | traL | 1302925..1303206 [+], 282 | type IV conjugative transfer system protein TraL | TraL_F, T4SS component |
| 40 | I636_RS05855 | 1303203..1303829 [+], 627 | hypothetical protein | TraE_F, T4SS component |
| 41 | I636_RS05860 | 1303813..1304709 [+], 897 | conjugal transfer protein TraK | TraK_F, T4SS component |
| 42 | I636_RS05865 | 1304712..1306001 [+], 1290 | IncF plasmid conjugative transfer pilus assembly protein TraB | TraB_F, T4SS component |
| 43 | I636_RS05870 | 1305998..1306648 [+], 651 | conjugal transfer protein TraV | TraV_F, T4SS component |
| 44 | I636_RS05875 | 1306645..1307034 [+], 390 | hypothetical protein | TraA_F, T4SS component |
| 45 | I636_RS05880 | 1307443..1309938 [+], 2496 | copper-translocating P-type ATPase | |
| 46 | cueR | 1309964..1310428 [+], 465 | Cu(I)-responsive transcriptional regulator | |
| 47 | I636_RS05895 | 1311214..1311405 [+], 192 | hypothetical protein | |
| 48 | cueR | 1312013..1312438 [+], 426 | Cu(I)-responsive transcriptional regulator | |
| 49 | I636_RS05905 | 1313201..1313554 [+], 354 | ArsR family transcriptional regulator | |
| 50 | I636_RS05915 | 1314913..1315941 [+], 1029 | cation transporter | |
| 51 | I636_RS05920 | 1315986..1316264 [+], 279 | transcriptional regulator | |
| 52 | I636_RS05925 | 1316861..1319515 [+], 2655 | cadmium-translocating P-type ATPase | |
| 53 | I636_RS05935 | 1321184..1324375 [+], 3192 | multidrug efflux RND transporter permease subunit | |
| 54 | I636_RS05940 | 1324377..1325870 [+], 1494 | multidrug transporter | |
| 55 | I636_RS05945 | 1326343..1326693 [+], 351 | AcrB/AcrD/AcrF family protein | |
| 56 | I636_RS19830 | 1326684..1326800 [+], 117 | AcrB/AcrD/AcrF family protein | |
| 57 | I636_RS05950 | 1326846..1327067 [+], 222 | hydrophobe/amphiphile efflux protein | |
| 58 | I636_RS19685 | 1327165..1327356 [+], 192 | copper chaperone | |
| 59 | I636_RS19690 | 1328057..1328329 [+], 273 | IS66 family insertion sequence hypothetical protein | |
| 60 | I636_RS05960 | 1328326..1328580 [+], 255 | IS66 family insertion sequence hypothetical protein | |
| 61 | I636_RS05965 | 1328791..1329483 [+], 693 | DsbC family protein | TrbB_I, T4SS component |
| 62 | traC | 1329483..1331882 [+], 2400 | type IV secretion system protein TraC | TraC_F, T4SS component |
| 63 | I636_RS05975 | 1331875..1332222 [+], 348 | hypothetical protein | |
| 64 | I636_RS05980 | 1332299..1332718 [+], 420 | conjugal transfer pilin signal peptidase TrbI | TraF, T4SS component |
| 65 | I636_RS05985 | 1332729..1333853 [+], 1125 | plasmid pilus assembly protein | TraW_F, T4SS component |
| 66 | I636_RS05990 | 1333837..1334865 [+], 1029 | TraU family protein | TraU_F, T4SS component |
| 67 | I636_RS05995 | 1334868..1338560 [+], 3693 | mating pair stabilization TraN | TraN_F, T4SS component |
| 68 | I636_RS06000 | 1338702..1340378 [-], 1677 | ATP-dependent helicase | |
| 69 | I636_RS06005 | 1340375..1342300 [-], 1926 | ATP-dependent endonuclease | |
| 70 | I636_RS06010 | 1342378..1344756 [-], 2379 | peptidase S8 | |
| 71 | I636_RS06015 | 1344771..1345742 [-], 972 | ATP-binding protein | |
| 72 | I636_RS06020 | 1345745..1345942 [-], 198 | hypothetical protein | |
| 73 | I636_RS06025 | 1346084..1346686 [-], 603 | hypothetical protein | |
| 74 | I636_RS06030 | 1347054..1347380 [+], 327 | hypothetical protein | |
| 75 | I636_RS06035 | 1347396..1347815 [+], 420 | single-stranded DNA-binding protein | |
| 76 | bet | 1347895..1348713 [+], 819 | phage recombination protein Bet | |
| 77 | I636_RS06045 | 1349000..1350016 [+], 1017 | DNA recombination protein | |
| 78 | I636_RS06050 | 1350226..1351185 [+], 960 | AAA family ATPase | |
| 79 | I636_RS06055 | 1351185..1351952 [+], 768 | hypothetical protein | |
| 80 | I636_RS06060 | 1352051..1353004 [+], 954 | DUF3150 domain-containing protein | |
| 81 | I636_RS06065 | 1353066..1353506 [+], 441 | hypothetical protein | |
| 82 | I636_RS06070 | 1353576..1355231 [+], 1656 | VWA domain-containing protein | |
| 83 | I636_RS06075 | 1355317..1355814 [+], 498 | DNA repair protein RadC | |
| 84 | I636_RS06080 | 1355814..1356155 [+], 342 | hypothetical protein | |
| 85 | I636_RS06085 | 1356247..1357314 [+], 1068 | phage P4 alpha, zinc-binding domain-containing protein | |
| 86 | I636_RS06090 | 1357404..1358111 [+], 708 | hypothetical protein | |
| 87 | I636_RS06095 | 1358354..1359271 [-], 918 | LysR family transcriptional regulator | |
| 88 | I636_RS06100 | 1359384..1360523 [+], 1140 | chromate resistance protein | |
| 89 | I636_RS06105 | 1360895..1361839 [+], 945 | thioredoxin family protein | TraF_F, T4SS component |
| 90 | I636_RS06110 | 1361842..1363230 [+], 1389 | TraH family protein | TraH_F, T4SS component |
| 91 | I636_RS06115 | 1363234..1366803 [+], 3570 | conjugal transfer protein TraG | TraG_F, T4SS component |
| 92 | I636_RS06120 | 1366836..1367267 [-], 432 | hypothetical protein | |
| 93 | I636_RS06125 | 1367325..1367858 [-], 534 | transcriptional activator | |
| 94 | I636_RS06130 | 1367855..1368154 [-], 300 | transcriptional activator | |
| 95 | I636_RS06135 | 1368151..1368699 [-], 549 | lytic transglycosylase domain-containing protein | Orf169_F, T4SS component |
| 96 | I636_RS06140 | 1368686..1369348 [-], 663 | hypothetical protein | |
| 97 | I636_RS06145 | 1369335..1370204 [-], 870 | hypothetical protein | |
| 98 | I636_RS06150 | 1370260..1370511 [-], 252 | hypothetical protein | |
| 99 | I636_RS06155 | 1370629..1371276 [+], 648 | LexA family transcriptional regulator | |
| 100 | prfC | 1371533..1373122 [+], 1590 | peptide chain release factor 3 | |
| 101 | I636_RS06165 | 1373136..1374866 [+], 1731 | arginine--tRNA ligase | |
| 102 | I636_RS06170 | 1375059..1375625 [+], 567 | NADPH-dependent oxidoreductase | |
| 103 | I636_RS06175 | 1375838..1376041 [+], 204 | hypothetical protein | |
| 104 | I636_RS06180 | 1376149..1377405 [-], 1257 | DUF2891 domain-containing protein | |
| 105 | I636_RS06185 | 1377465..1378508 [-], 1044 | PLP-dependent cysteine synthase family protein | |
| 106 | I636_RS06190 | 1378835..1380049 [+], 1215 | endonuclease/exonuclease/phosphatase family protein | |
| 107 | I636_RS06195 | 1380167..1383010 [+], 2844 | GGDEF domain-containing protein | |