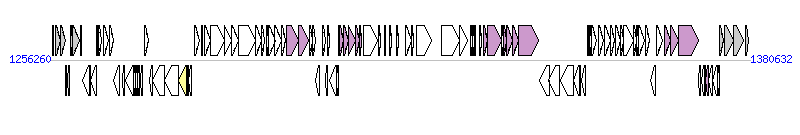The graph information of ICEAmaAnS1 components from NZ_CP014323 |
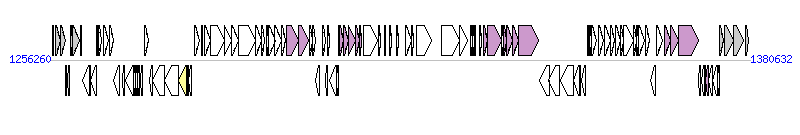 |
| Incomplete gene list of ICEAmaAnS1 from NZ_CP014323 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | AVL55_RS20225 | 1256716..1256877 [+], 162 | antirestriction protein family protein | |
| 2 | AVL55_RS05585 | 1257198..1258049 [+], 852 | hypothetical protein | |
| 3 | AVL55_RS05590 | 1258042..1258965 [+], 924 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | |
| 4 | AVL55_RS05595 | 1258980..1259183 [-], 204 | hypothetical protein | |
| 5 | AVL55_RS05600 | 1259447..1259635 [-], 189 | hypothetical protein | |
| 6 | AVL55_RS05605 | 1259986..1260306 [+], 321 | XRE family transcriptional regulator | |
| 7 | AVL55_RS05610 | 1260299..1261528 [+], 1230 | type II toxin-antitoxin system HipA family toxin | |
| 8 | AVL55_RS05615 | 1261745..1261939 [+], 195 | DNA-binding protein | |
| 9 | AVL55_RS05620 | 1261951..1263192 [-], 1242 | DUF4102 domain-containing protein | |
| 10 | AVL55_RS05625 | 1263194..1263463 [-], 270 | hypothetical protein | |
| 11 | AVL55_RS05630 | 1263466..1264440 [-], 975 | rod shape determination protein | |
| 12 | AVL55_RS20230 | 1264452..1264658 [+], 207 | hypothetical protein | |
| 13 | AVL55_RS05635 | 1264905..1265351 [+], 447 | hypothetical protein | |
| 14 | AVL55_RS05640 | 1265716..1266684 [+], 969 | calcium:sodium antiporter | |
| 15 | AVL55_RS05645 | 1266753..1267586 [+], 834 | mechanosensitive ion channel family protein | |
| 16 | AVL55_RS05650 | 1267595..1268605 [-], 1011 | translesion error-prone DNA polymerase V subunit UmuC | |
| 17 | AVL55_RS05655 | 1269091..1269528 [-], 438 | MerR family transcriptional regulator | |
| 18 | AVL55_RS05660 | 1269642..1271081 [-], 1440 | mercury(II) reductase | |
| 19 | AVL55_RS05665 | 1271102..1271521 [-], 420 | mercury transporter MerC | |
| 20 | AVL55_RS05670 | 1271521..1271805 [-], 285 | mercury resistance system periplasmic binding protein MerP | |
| 21 | AVL55_RS05675 | 1271852..1272199 [-], 348 | mercury transporter | |
| 22 | AVL55_RS05680 | 1272390..1272761 [-], 372 | Hg(II)-responsive transcriptional regulator | |
| 23 | AVL55_RS05685 | 1273112..1273735 [+], 624 | serine recombinase | |
| 24 | AVL55_RS05690 | 1273858..1274394 [-], 537 | hypothetical protein | |
| 25 | AVL55_RS05695 | 1274398..1276587 [-], 2190 | hypothetical protein | |
| 26 | AVL55_RS05700 | 1276599..1279067 [-], 2469 | hypothetical protein | |
| 27 | AVL55_RS05705 | 1279072..1280481 [-], 1410 | site-specific integrase | Integrase |
| 28 | AVL55_RS05710 | 1280634..1280903 [-], 270 | DNA polymerase | |
| 29 | AVL55_RS05715 | 1280911..1281360 [-], 450 | translesion error-prone DNA polymerase V autoproteolytic subunit | |
| 30 | AVL55_RS05720 | 1282000..1282905 [+], 906 | DNA polymerase III subunit epsilon | |
| 31 | AVL55_RS05725 | 1283119..1283418 [+], 300 | hypothetical protein | |
| 32 | AVL55_RS05730 | 1283736..1284704 [+], 969 | WYL domain-containing protein | |
| 33 | AVL55_RS05735 | 1284818..1287247 [+], 2430 | SAM-dependent DNA methyltransferase | |
| 34 | AVL55_RS05740 | 1287247..1288515 [+], 1269 | restriction endonuclease subunit S | |
| 35 | AVL55_RS05745 | 1288525..1289688 [+], 1164 | DUF4268 domain-containing protein | |
| 36 | AVL55_RS05750 | 1289703..1292720 [+], 3018 | type I restriction endonuclease subunit R | |
| 37 | AVL55_RS05755 | 1292743..1293828 [+], 1086 | ATP-binding region, ATPase-like protein | |
| 38 | AVL55_RS05760 | 1293851..1294402 [+], 552 | MarR family transcriptional regulator | |
| 39 | AVL55_RS05765 | 1294707..1295075 [+], 369 | metal-dependent hydrolase | |
| 40 | AVL55_RS05770 | 1295085..1296239 [+], 1155 | SAVED domain-containing protein | |
| 41 | AVL55_RS05775 | 1296232..1297422 [+], 1191 | nucleotidyltransferase | |
| 42 | AVL55_RS05780 | 1297423..1298136 [+], 714 | hypothetical protein | |
| 43 | AVL55_RS05785 | 1298253..1300403 [+], 2151 | metal-dependent phosphohydrolase | Relaxase, MOBH Family |
| 44 | AVL55_RS05790 | 1300452..1302275 [+], 1824 | hypothetical protein | TraD_F, T4SS component |
| 45 | AVL55_RS05795 | 1302285..1302845 [+], 561 | hypothetical protein | |
| 46 | AVL55_RS05800 | 1302832..1303467 [+], 636 | DUF4400 domain-containing protein | |
| 47 | AVL55_RS05805 | 1303494..1304081 [-], 588 | hypothetical protein | |
| 48 | AVL55_RS05810 | 1304700..1305122 [+], 423 | hypothetical protein | |
| 49 | AVL55_RS05815 | 1305176..1305502 [-], 327 | sel1 repeat family protein | |
| 50 | AVL55_RS05820 | 1305545..1305868 [+], 324 | hypothetical protein | |
| 51 | AVL55_RS05825 | 1305962..1307125 [-], 1164 | hypothetical protein | |
| 52 | AVL55_RS05830 | 1307250..1307462 [-], 213 | hypothetical protein | |
| 53 | AVL55_RS05835 | 1307498..1307779 [+], 282 | type IV conjugative transfer system protein TraL | TraL_F, T4SS component |
| 54 | AVL55_RS05840 | 1307776..1308402 [+], 627 | hypothetical protein | TraE_F, T4SS component |
| 55 | AVL55_RS05845 | 1308386..1309282 [+], 897 | hypothetical protein | TraK_F, T4SS component |
| 56 | AVL55_RS05850 | 1309285..1310574 [+], 1290 | conjugal transfer protein TraB | TraB_F, T4SS component |
| 57 | AVL55_RS05855 | 1310571..1311221 [+], 651 | conjugal transfer protein TraV | TraV_F, T4SS component |
| 58 | AVL55_RS05860 | 1311218..1311607 [+], 390 | hypothetical protein | TraA_F, T4SS component |
| 59 | AVL55_RS05865 | 1312016..1314511 [+], 2496 | copper-translocating P-type ATPase | |
| 60 | AVL55_RS05870 | 1314537..1315001 [+], 465 | Cu(I)-responsive transcriptional regulator | |
| 61 | AVL55_RS05880 | 1315684..1315902 [+], 219 | hypothetical protein | |
| 62 | AVL55_RS05885 | 1316587..1317012 [+], 426 | Cu(I)-responsive transcriptional regulator | |
| 63 | AVL55_RS05890 | 1317775..1318128 [+], 354 | ArsR family transcriptional regulator | |
| 64 | AVL55_RS05900 | 1319481..1320509 [+], 1029 | cation transporter | |
| 65 | AVL55_RS05905 | 1320554..1320832 [+], 279 | transcriptional regulator | |
| 66 | AVL55_RS05910 | 1321429..1324083 [+], 2655 | cadmium-translocating P-type ATPase | |
| 67 | AVL55_RS05920 | 1325752..1328943 [+], 3192 | multidrug efflux RND transporter permease subunit | |
| 68 | AVL55_RS05925 | 1328945..1330438 [+], 1494 | multidrug transporter | |
| 69 | AVL55_RS05930 | 1330950..1331261 [+], 312 | AcrB/AcrD/AcrF family protein | |
| 70 | AVL55_RS05935 | 1331414..1331635 [+], 222 | hydrophobe/amphiphile efflux protein | |
| 71 | AVL55_RS20255 | 1331733..1331924 [+], 192 | copper chaperone | |
| 72 | AVL55_RS20260 | 1332625..1332897 [+], 273 | IS66 family insertion sequence hypothetical protein | |
| 73 | AVL55_RS05950 | 1333360..1334052 [+], 693 | DsbC family protein | TrbB_I, T4SS component |
| 74 | AVL55_RS05955 | 1334052..1336451 [+], 2400 | type IV secretion system protein TraC | TraC_F, T4SS component |
| 75 | AVL55_RS05960 | 1336444..1336791 [+], 348 | hypothetical protein | |
| 76 | AVL55_RS05965 | 1336868..1337287 [+], 420 | conjugal transfer pilin signal peptidase TrbI | TraF, T4SS component |
| 77 | AVL55_RS05970 | 1337298..1338422 [+], 1125 | plasmid pilus assembly protein | TraW_F, T4SS component |
| 78 | AVL55_RS05975 | 1338406..1339434 [+], 1029 | TraU family protein | TraU_F, T4SS component |
| 79 | AVL55_RS05980 | 1339437..1343102 [+], 3666 | conjugal transfer protein TraN | TraN_F, T4SS component |
| 80 | AVL55_RS05985 | 1343244..1344920 [-], 1677 | ATP-dependent helicase | |
| 81 | AVL55_RS05990 | 1344917..1346842 [-], 1926 | ATP-dependent endonuclease | |
| 82 | AVL55_RS05995 | 1346920..1349298 [-], 2379 | peptidase S8 | |
| 83 | AVL55_RS06000 | 1349313..1350284 [-], 972 | ATP-binding protein | |
| 84 | AVL55_RS06005 | 1350395..1350574 [-], 180 | hypothetical protein | |
| 85 | AVL55_RS06010 | 1350716..1351318 [-], 603 | hypothetical protein | |
| 86 | AVL55_RS06015 | 1351686..1352012 [+], 327 | hypothetical protein | |
| 87 | AVL55_RS06020 | 1352028..1352447 [+], 420 | single-stranded DNA-binding protein | |
| 88 | AVL55_RS06025 | 1352527..1353345 [+], 819 | phage recombination protein Bet | |
| 89 | AVL55_RS06030 | 1353632..1354648 [+], 1017 | DNA recombination protein | |
| 90 | AVL55_RS06035 | 1354858..1355817 [+], 960 | AAA family ATPase | |
| 91 | AVL55_RS06040 | 1355817..1356584 [+], 768 | hypothetical protein | |
| 92 | AVL55_RS06045 | 1356683..1357636 [+], 954 | DUF3150 domain-containing protein | |
| 93 | AVL55_RS06050 | 1357698..1358138 [+], 441 | hypothetical protein | |
| 94 | AVL55_RS06055 | 1358208..1359863 [+], 1656 | VWA domain-containing protein | |
| 95 | AVL55_RS06060 | 1359949..1360446 [+], 498 | DNA repair protein RadC | |
| 96 | AVL55_RS06065 | 1360446..1360787 [+], 342 | hypothetical protein | |
| 97 | AVL55_RS06070 | 1360879..1361946 [+], 1068 | phage P4 alpha, zinc-binding domain-containing protein | |
| 98 | AVL55_RS06075 | 1362036..1362743 [+], 708 | hypothetical protein | |
| 99 | AVL55_RS06080 | 1362986..1363927 [-], 942 | LysR family transcriptional regulator | |
| 100 | AVL55_RS06085 | 1364016..1365155 [+], 1140 | chromate resistance protein | |
| 101 | AVL55_RS06090 | 1365527..1366471 [+], 945 | thioredoxin family protein | TraF_F, T4SS component |
| 102 | AVL55_RS06095 | 1366474..1367862 [+], 1389 | TraH family protein | TraH_F, T4SS component |
| 103 | AVL55_RS06100 | 1367866..1371435 [+], 3570 | conjugal transfer protein TraG | TraG_F, T4SS component |
| 104 | AVL55_RS06105 | 1371468..1371899 [-], 432 | hypothetical protein | |
| 105 | AVL55_RS06110 | 1371957..1372490 [-], 534 | transcriptional activator | |
| 106 | AVL55_RS06115 | 1372487..1372786 [-], 300 | transcriptional activator | |
| 107 | AVL55_RS06120 | 1372783..1373331 [-], 549 | lytic transglycosylase domain-containing protein | Orf169_F, T4SS component |
| 108 | AVL55_RS06125 | 1373318..1373980 [-], 663 | hypothetical protein | |
| 109 | AVL55_RS06130 | 1373967..1374836 [-], 870 | hypothetical protein | |
| 110 | AVL55_RS06135 | 1374892..1375143 [-], 252 | hypothetical protein | |
| 111 | AVL55_RS06140 | 1375261..1375908 [+], 648 | LexA family transcriptional regulator | |
| 112 | AVL55_RS06145 | 1376165..1377754 [+], 1590 | peptide chain release factor 3 | |
| 113 | AVL55_RS06150 | 1377788..1379518 [+], 1731 | arginine--tRNA ligase | |
| 114 | AVL55_RS06155 | 1379807..1380373 [+], 567 | NADPH-dependent oxidoreductase | |