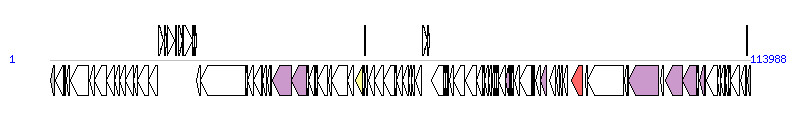The graph information of ICESsuZJ20091101-2 components from KX077883 |
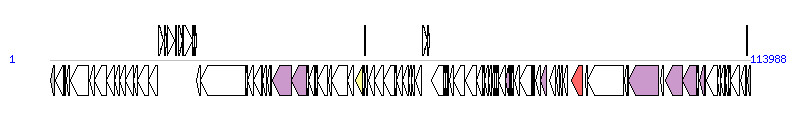 |
| Complete gene list of ICESsuZJ20091101-2 from KX077883 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | hyd | 1..750 [-], 750 | putative hydrolase (HAD superfamily) | |
| 2 | - | 937..2145 [-], 1209 | integrase | |
| 3 | - | 2138..2386 [-], 249 | hypothetical protein | |
| 4 | - | 2470..2598 [-], 129 | hypothetical protein | |
| 5 | - | 2722..3138 [-], 417 | hypothetical protein | |
| 6 | - | 3346..6285 [-], 2940 | fucolectin-related protein | |
| 7 | - | 6428..7321 [-], 894 | hexokinase | |
| 8 | - | 7323..9200 [-], 1878 | enterotoxin | |
| 9 | - | 9190..10524 [-], 1335 | alpha-L-fucosidase | |
| 10 | - | 10521..11360 [-], 840 | multiple sugar ABC transporter, membrane-spanning | |
| 11 | - | 11371..12291 [-], 921 | N-Acetyl-D-glucosamine ABC transport system | |
| 12 | - | 12346..13644 [-], 1299 | N-Acetyl-D-glucosamine ABC transport system | |
| 13 | - | 13661..14305 [-], 645 | L-fuculose phosphate aldolase | |
| 14 | - | 14320..16107 [-], 1788 | L-fucose isomerase | |
| 15 | - | 16091..17518 [-], 1428 | L-fuculokinase | |
| 16 | - | 17677..18447 [+], 771 | L-fucose operon activator | |
| 17 | - | 18621..18974 [+], 354 | hypothetical protein | |
| 18 | - | 19081..20271 [+], 1191 | Rlx-like protein | |
| 19 | - | 20434..20592 [+], 159 | hypothetical protein | |
| 20 | - | 20937..21629 [+], 693 | pleiotropic regulator of exopolysaccharide | |
| 21 | - | 21632..21766 [+], 135 | hypothetical protein | |
| 22 | - | 21772..23187 [+], 1416 | ImpB/MucB/SamB family protein | |
| 23 | - | 23184..23534 [+], 351 | hypothetical protein | |
| 24 | - | 23521..23814 [+], 294 | hypothetical protein | |
| 25 | - | 23861..24499 [-], 639 | hypothetical protein | |
| 26 | - | 24496..31767 [-], 7272 | superfamily II DNA and RNA helicase | |
| 27 | - | 31786..32238 [-], 453 | hypothetical protein | |
| 28 | - | 32235..33194 [-], 960 | modification methylase ScrFIB | |
| 29 | - | 33191..34375 [-], 1185 | DNA-cytosine methyltransferase | |
| 30 | - | 34452..34682 [-], 231 | hypothetical protein | |
| 31 | - | 34667..35305 [-], 639 | hypothetical protein | |
| 32 | - | 35348..35944 [-], 597 | mature parasite-infected erythrocyte surface | |
| 33 | - | 35928..36143 [-], 216 | hypothetical protein | |
| 34 | - | 36154..39375 [-], 3222 | immunogenic secreted protein | Orf13_p, T4SS component |
| 35 | - | 39383..41812 [-], 2430 | TrsE-like protein | T4CP |
| 36 | - | 41769..42137 [-], 369 | hypothetical protein | |
| 37 | - | 42150..43013 [-], 864 | hypothetical protein | |
| 38 | - | 43022..43237 [-], 216 | hypothetical protein | |
| 39 | - | 43237..43545 [-], 309 | hypothetical protein | |
| 40 | - | 43621..45213 [-], 1593 | TrsK-like protein | |
| 41 | - | 45420..45914 [-], 495 | hypothetical protein | |
| 42 | - | 45907..48501 [-], 2595 | virulence-associated cell-wall-anchored protein | |
| 43 | - | 48498..49457 [-], 960 | hypothetical protein | |
| 44 | - | 49718..50923 [-], 1206 | integrase | Integrase |
| 45 | - | 50926..51174 [-], 249 | hypothetical protein | |
| 46 | - | 51233..51376 [+], 144 | hypothetical protein | |
| 47 | - | 51444..51668 [-], 225 | putative repressor protein | |
| 48 | - | 51915..52811 [-], 897 | LacX protein | |
| 49 | - | 52899..54305 [-], 1407 | 6-phospho-beta-galactosidase | |
| 50 | - | 54371..56074 [-], 1704 | PTS system, lactose-specific IIB component | |
| 51 | - | 56074..56391 [-], 318 | PTS system, lactose-specific IIA component | |
| 52 | - | 56419..57399 [-], 981 | tagatose 1,6-bisphosphate aldolase | |
| 53 | - | 57402..58334 [-], 933 | tagatose-6-phosphate kinase | |
| 54 | - | 58346..58861 [-], 516 | galactose-6-phosphate isomerase LacB subunit | |
| 55 | - | 58895..59320 [-], 426 | galactose-6-phosphate isomerase LacA subunit | |
| 56 | - | 59558..60508 [-], 951 | glucokinase | |
| 57 | - | 60688..61449 [+], 762 | lactose phosphotransferase system repressor | |
| 58 | - | 61513..61866 [+], 354 | putative FMN hydrolase | |
| 59 | - | 62169..64034 [-], 1866 | relaxase | |
| 60 | - | 64021..64386 [-], 366 | hypothetical protein | |
| 61 | - | 64396..64755 [-], 360 | hypothetical protein | |
| 62 | - | 64849..64971 [-], 123 | hypothetical protein | |
| 63 | - | 65081..65665 [-], 585 | hypothetical protein | |
| 64 | - | 65712..67505 [-], 1794 | ATP-dependent DNA helicase | |
| 65 | - | 67634..69463 [-], 1830 | hypothetical protein | |
| 66 | - | 69465..70409 [-], 945 | mobile element protein | |
| 67 | - | 70494..70703 [-], 210 | hypothetical protein | |
| 68 | - | 70684..71478 [-], 795 | zeta toxin | |
| 69 | - | 71478..71954 [-], 477 | transcription regulator, probable | |
| 70 | - | 72024..72308 [-], 285 | hypothetical protein | |
| 71 | - | 72352..72747 [-], 396 | methyl-accepting chemotaxis protein | |
| 72 | - | 72751..72978 [-], 228 | hypothetical protein | |
| 73 | - | 73032..74120 [-], 1089 | hypothetical protein | |
| 74 | - | 74160..74798 [-], 639 | hypothetical protein | PrgL, T4SS component |
| 75 | - | 74788..74904 [-], 117 | hypothetical protein | |
| 76 | - | 74909..75199 [-], 291 | hypothetical protein | |
| 77 | - | 75214..75513 [-], 300 | hypothetical protein | |
| 78 | - | 75583..76494 [-], 912 | SNF2 family protein | |
| 79 | - | 76509..78392 [-], 1884 | site-specific recombinase | |
| 80 | - | 78446..78637 [-], 192 | hypothetical protein | |
| 81 | - | 78708..78938 [-], 231 | bacterial seryl-tRNA synthetase related protein | |
| 82 | - | 79015..79869 [-], 855 | chromosome (plasmid) partitioning protein ParB | |
| 83 | - | 79877..80842 [-], 966 | hypothetical protein | Relaxase, MOBV Family |
| 84 | - | 81361..82392 [-], 1032 | hypothetical protein | |
| 85 | - | 82546..83019 [-], 474 | RNA polymerase ECF-type sigma factor | |
| 86 | - | 83103..83528 [-], 426 | hypothetical protein | |
| 87 | - | 83623..84258 [-], 636 | hypothetical protein | |
| 88 | - | 84847..86766 [-], 1920 | tetracycline resistance protein TetO | AR |
| 89 | - | 87133..87507 [-], 375 | TnpV | |
| 90 | - | 87488..93421 [-], 5934 | SNF2 family protein | |
| 91 | - | 93472..94023 [-], 552 | hypothetical protein | |
| 92 | - | 94007..94198 [-], 192 | putative calcium-binding protein | |
| 93 | - | 94199..99103 [-], 4905 | agglutinin receptor | PrgB, T4SS component |
| 94 | - | 99345..99938 [-], 594 | hypothetical protein | |
| 95 | - | 100183..102987 [-], 2805 | hypothetical protein | PrgK, T4SS component |
| 96 | - | 102984..105314 [-], 2331 | TrsE-like protein | PrgJ, T4SS component |
| 97 | - | 105292..105645 [-], 354 | hypothetical protein | |
| 98 | - | 105708..106562 [-], 855 | hypothetical protein | PrgH, T4SS component |
| 99 | - | 106579..106821 [-], 243 | hypothetical protein | |
| 100 | - | 106839..108659 [-], 1821 | hypothetical protein | |
| 101 | - | 108659..109147 [-], 489 | hypothetical protein | |
| 102 | - | 109224..109805 [-], 582 | hypothetical protein | |
| 103 | - | 109808..110041 [-], 234 | hypothetical protein | |
| 104 | - | 110051..110434 [-], 384 | hypothetical protein | |
| 105 | - | 110444..110872 [-], 429 | hypothetical protein | |
| 106 | - | 110856..112211 [-], 1356 | modification methylase | |
| 107 | - | 112369..113193 [-], 825 | hypothetical protein | |
| 108 | - | 113190..113363 [-], 174 | hypothetical protein | |
| 109 | - | 113447..113560 [+], 114 | hypothetical protein | |
| 110 | - | 113623..113988 [-], 366 | LSU ribosomal protein L7/L12 (P1/P2) | |