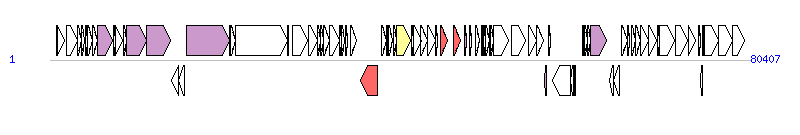The graph information of ICESsuJH1301 components from KX077886 |
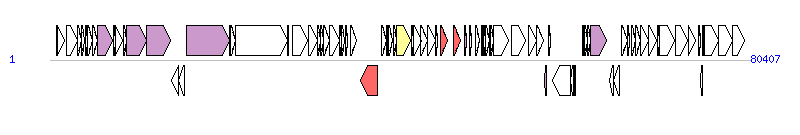 |
| Complete gene list of ICESsuJH1301 from KX077886 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 717..884 [+], 168 | hypothetical protein | |
| 2 | - | 887..1705 [+], 819 | hypothetical protein | |
| 3 | - | 1854..3209 [+], 1356 | DNA-cytosine methyltransferase | |
| 4 | - | 3193..3621 [+], 429 | hypothetical protein | |
| 5 | - | 3632..4015 [+], 384 | hypothetical protein | |
| 6 | - | 4024..4257 [+], 234 | hypothetical protein | |
| 7 | - | 4260..4844 [+], 585 | hypothetical protein | |
| 8 | - | 4921..5409 [+], 489 | hypothetical protein | |
| 9 | - | 5409..7226 [+], 1818 | hypothetical protein | T4CP |
| 10 | - | 7244..7486 [+], 243 | hypothetical protein | |
| 11 | - | 7505..8359 [+], 855 | hypothetical protein | |
| 12 | - | 8421..8774 [+], 354 | hypothetical protein | |
| 13 | - | 8731..11082 [+], 2352 | TrsE-like protein | PrgJ, T4SS component |
| 14 | - | 11084..13885 [+], 2802 | hypothetical protein | PrgK, T4SS component |
| 15 | - | 13972..14817 [-], 846 | abortive infection protein AbiGII | |
| 16 | - | 14814..15404 [-], 591 | abortive infection protein AbiGI | |
| 17 | - | 15685..20580 [+], 4896 | agglutinin receptor | PrgB, T4SS component |
| 18 | - | 20581..20772 [+], 192 | putative calcium-binding protein | |
| 19 | - | 20756..21307 [+], 552 | hypothetical protein | |
| 20 | - | 21356..27259 [+], 5904 | SNF2 family protein | |
| 21 | - | 27289..27402 [+], 114 | hypothetical protein | |
| 22 | - | 27824..29635 [+], 1812 | tetron-type RNA-directed DNA polymerase | |
| 23 | - | 29695..30687 [+], 993 | SNF2 family protein | |
| 24 | - | 30758..31057 [+], 300 | hypothetical protein | |
| 25 | - | 31071..31361 [+], 291 | hypothetical protein | |
| 26 | - | 31457..32095 [+], 639 | hypothetical protein | |
| 27 | - | 32135..33211 [+], 1077 | hypothetical protein | |
| 28 | - | 33265..33492 [+], 228 | hypothetical protein | |
| 29 | - | 33489..33878 [+], 390 | methyl-accepting chemotaxis protein | |
| 30 | - | 33928..34224 [+], 297 | hypothetical protein | |
| 31 | - | 34513..35190 [+], 678 | transposase | |
| 32 | - | 35683..37608 [-], 1926 | ribosome protection-type tetracycline resistance | AR |
| 33 | - | 38124..38606 [+], 483 | DNA topoisomerase III | |
| 34 | - | 38678..38794 [+], 117 | DNA topoisomerase III | |
| 35 | - | 38902..39399 [+], 498 | putative DNA recombinase | |
| 36 | - | 39400..39816 [+], 417 | recombinase | |
| 37 | - | 39818..41380 [+], 1563 | putative DNA recombinase | Integrase |
| 38 | - | 41523..41747 [+], 225 | helix-turn-helix domain protein | |
| 39 | - | 41762..42631 [+], 870 | hypothetical protein | |
| 40 | - | 42612..43346 [+], 735 | ubiquinone/menaquinone biosynthesis | |
| 41 | - | 43379..44287 [+], 909 | putative aminoglycoside 6-adenylyltansferase | |
| 42 | - | 44284..44538 [+], 255 | streptothricin acetyltransferase | |
| 43 | - | 44857..45651 [+], 795 | aminoglycoside phosphotransferase | AR |
| 44 | - | 46400..47137 [+], 738 | rRNA (adenine-N(6)-) -methyltransferase | AR |
| 45 | - | 47620..47859 [+], 240 | pheromone response surface protein PrgC | Orf15_p, T4SS component |
| 46 | - | 48181..48459 [+], 279 | CopS protein | |
| 47 | - | 48856..49386 [+], 531 | mobile element protein | |
| 48 | - | 49578..49709 [+], 132 | arsenical resistance operon repressor | |
| 49 | - | 49769..50062 [+], 294 | arsenical resistance operon repressor | |
| 50 | - | 50064..50435 [+], 372 | arsenical resistance operon trans-acting | |
| 51 | - | 50455..50580 [+], 126 | hypothetical protein | |
| 52 | - | 50570..50917 [+], 348 | arsenical resistance operon repressor | |
| 53 | - | 50953..52695 [+], 1743 | arsenical pump-driving ATPase | |
| 54 | - | 52984..54657 [+], 1674 | pyridine nucleotide-disulfide oxidoreductase | |
| 55 | - | 54947..55882 [+], 936 | ABC transporter, ATP-binding protein | |
| 56 | - | 55875..56645 [+], 771 | ABC transporter, permease protein | |
| 57 | - | 56758..56988 [-], 231 | putative secreted cell wall protein | YddH, T4SS component |
| 58 | - | 57246..57464 [+], 219 | hypothetical protein | |
| 59 | - | 57698..59815 [-], 2118 | cadmium-transporting ATPase | |
| 60 | - | 59812..60171 [-], 360 | cadmium efflux system accessory protein | |
| 61 | - | 60255..60368 [-], 114 | hypothetical protein | |
| 62 | - | 61201..61338 [+], 138 | hypothetical protein | |
| 63 | - | 61357..61716 [+], 360 | hypothetical protein | |
| 64 | - | 61726..62091 [+], 366 | hypothetical protein | |
| 65 | - | 62078..63943 [+], 1866 | relaxase | Relaxase, MOBP Family |
| 66 | - | 64274..64681 [-], 408 | putative FMN hydrolase | |
| 67 | - | 64690..65451 [-], 762 | lactose phosphotransferase system repressor | |
| 68 | - | 65632..66312 [+], 681 | glucokinase | |
| 69 | - | 66379..66582 [+], 204 | glucokinase | |
| 70 | - | 66816..67241 [+], 426 | galactose-6-phosphate isomerase LacA subunit | |
| 71 | - | 67269..67784 [+], 516 | galactose-6-phosphate isomerase LacB subunit | |
| 72 | - | 67796..68728 [+], 933 | tagatose-6-phosphate kinase | |
| 73 | - | 68731..69711 [+], 981 | tagatose 1,6-bisphosphate aldolase | |
| 74 | - | 69739..70056 [+], 318 | PTS system, lactose-specific IIA component | |
| 75 | - | 70056..71741 [+], 1686 | PTS system, lactose-specific IIB component | |
| 76 | - | 71807..73213 [+], 1407 | 6-phospho-beta-galactosidase | |
| 77 | - | 73301..74197 [+], 897 | LacX protein | |
| 78 | - | 74444..74668 [+], 225 | putative repressor protein | |
| 79 | - | 74736..74894 [-], 159 | hypothetical protein | |
| 80 | - | 74940..75080 [+], 141 | hypothetical protein | |
| 81 | - | 75179..76753 [+], 1575 | site-specific recombinase | |
| 82 | - | 76753..78372 [+], 1620 | site-specific recombinase | |
| 83 | - | 78365..79813 [+], 1449 | site-specific recombinase | |