The graph information of ICESpy009 components from KU056701 |
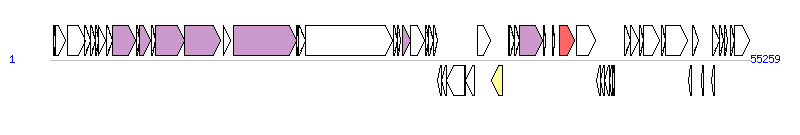 |
| Complete gene list of ICESpy009 from KU056701 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 264..437 [+], 174 | hypothetical protein | |
| 2 | rep | 434..1258 [+], 825 | Replication initiator protein | |
| 3 | - | 1416..2771 [+], 1356 | DNA-cytosine methyltransferase | |
| 4 | - | 2755..3183 [+], 429 | hypothetycal protein | |
| 5 | - | 3193..3576 [+], 384 | ArsC family protein | |
| 6 | - | 3586..3819 [+], 234 | hypothetical protein | |
| 7 | - | 3822..4403 [+], 582 | hypothetical protein | |
| 8 | - | 4480..4968 [+], 489 | hypothetical protein | |
| 9 | virD4 | 4968..6788 [+], 1821 | putative Type IV secretion protein VirD4 | T4CP |
| 10 | - | 6806..7048 [+], 243 | membrane protein | |
| 11 | virB6 | 7065..7919 [+], 855 | putative Type IV secretion protein VirB6 | PrgH, T4SS component |
| 12 | - | 7981..8334 [+], 354 | hypothetical protein | |
| 13 | virB4 | 8291..10642 [+], 2352 | putative Type IV secretion protein VirB4 | PrgJ, T4SS component |
| 14 | virB1-like | 10639..13443 [+], 2805 | putative Type IV secretion protein VirB1-like protein | PrgK, T4SS component |
| 15 | - | 13688..14281 [+], 594 | hypothetical protein | |
| 16 | - | 14525..19429 [+], 4905 | glucan binding protein | PrgB, T4SS component |
| 17 | - | 19430..19621 [+], 192 | calcium binding protein | |
| 18 | - | 19605..20156 [+], 552 | hypothetical protein | |
| 19 | - | 20206..27027 [+], 6822 | SNF2 family protein | |
| 20 | - | 27097..27396 [+], 300 | hypothetical protein | |
| 21 | - | 27411..27701 [+], 291 | membrane protein | |
| 22 | - | 27812..28450 [+], 639 | hypothetical protein | PrgL, T4SS component |
| 23 | - | 28490..29578 [+], 1089 | DNA primase | |
| 24 | - | 29632..29847 [+], 216 | hypothetical protein | |
| 25 | - | 29863..30258 [+], 396 | chemotaxis protein | |
| 26 | - | 30302..30589 [+], 288 | ATPase | |
| 27 | - | 30616..30915 [-], 300 | hypothetical protein | |
| 28 | - | 30902..31270 [-], 369 | hypothetical protein | |
| 29 | - | 31267..32685 [-], 1419 | DNA-directed DNA polymerase | |
| 30 | - | 32687..32821 [-], 135 | hypothetical protein | |
| 31 | - | 32824..33516 [-], 693 | DNA binding protein | |
| 32 | - | 33730..34800 [+], 1071 | DNA recombination protein | |
| 33 | - | 34833..35687 [-], 855 | transposase | Integrase |
| 34 | - | 36172..36285 [+], 114 | hypothetical protein | |
| 35 | - | 36304..36663 [+], 360 | hypothetical protein | |
| 36 | - | 36673..37038 [+], 366 | hypothetical protein | |
| 37 | - | 37025..38887 [+], 1863 | relaxase | Relaxase, MOBP Family |
| 38 | - | 38923..39084 [+], 162 | hypothetical protein | |
| 39 | - | 39653..39838 [+], 186 | hypothetical protein | |
| 40 | mef(E) | 40224..41441 [+], 1218 | Macrolide efflux protein E | AR |
| 41 | mel | 41561..43024 [+], 1464 | Mel protein | |
| 42 | - | 43142..43441 [-], 300 | hypothetical protein | |
| 43 | - | 43428..43796 [-], 369 | hypothetical protein | |
| 44 | - | 43809..44144 [-], 336 | hypothetical protein | |
| 45 | - | 44141..44386 [-], 246 | ImpB/MucB/SamB family protein | |
| 46 | - | 44383..44571 [-], 189 | hypothetical protein | |
| 47 | - | 45351..45821 [+], 471 | transcriptional regulator | |
| 48 | - | 45805..46494 [+], 690 | putative aminoglycoside phosphotransferase | |
| 49 | - | 46498..46830 [+], 333 | putative aminoglycoside phosphotransferase | |
| 50 | - | 46827..48119 [+], 1293 | putative ABC transporter | |
| 51 | - | 48258..48572 [+], 315 | putative ABC transporter | |
| 52 | - | 48553..50292 [+], 1740 | putative ABC transporter | |
| 53 | - | 50404..50625 [-], 222 | putative transposase | |
| 54 | - | 50689..51183 [+], 495 | putative peptidase | |
| 55 | - | 51439..51606 [-], 168 | hypothetical protein | |
| 56 | - | 52222..52437 [-], 216 | hypothetical protein | |
| 57 | - | 52334..52843 [+], 510 | transposase | |
| 58 | - | 52840..53154 [+], 315 | transposase | |
| 59 | - | 53265..53483 [+], 219 | Cro/CI family transcriptional regulator | |
| 60 | - | 53687..54001 [+], 315 | hypothetical protein | |
| 61 | int | 54004..55209 [+], 1206 | integrase | |
