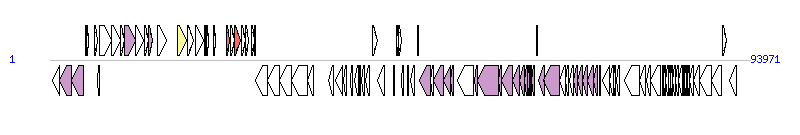The graph information of ICEEc2 components from GU725392 |
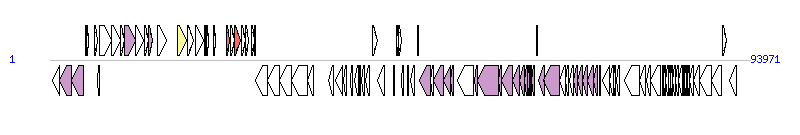 |
| Complete gene list of ICEEc2 from GU725392 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | ECAGIv1_0001 | 273..1262 [-], 990 | tyrosine recombinase xerC-like protein | |
| 2 | ECAGIv1_0002 | 1316..2938 [-], 1623 | hypothetical protein | Relaxase, MOBH Family |
| 3 | ECAGIv1_0003 | 2931..4439 [-], 1509 | putative superfamily I DNA and RNA helicase | TraI_F, T4SS component |
| 4 | ECAGIv1_0004 | 4728..4919 [+], 192 | hypothetical protein | |
| 5 | ECAGIv1_0005 | 4966..5184 [+], 219 | hypothetical protein | |
| 6 | ECAGIv1_0007 | 5926..6402 [+], 477 | putative PilM protein | |
| 7 | ECAGIv1_0008 | 6410..6622 [-], 213 | hypothetical protein | |
| 8 | pilN | 6610..8217 [+], 1608 | PilN | |
| 9 | pilO | 8241..9527 [+], 1287 | PilO | |
| 10 | pilP | 9517..9978 [+], 462 | pilus type IV assembly protein | |
| 11 | pilQ | 9988..11508 [+], 1521 | type IV pilus | TraJ_I, T4SS component |
| 12 | pilR | 11510..12610 [+], 1101 | PilR | |
| 13 | pilS | 12660..13196 [+], 537 | PilS | |
| 14 | pilT | 13255..13731 [+], 477 | PilT | Orf169_F, T4SS component |
| 15 | pilV | 14391..15683 [+], 1293 | PilV | |
| 16 | rci | 17170..18267 [+], 1098 | Rci | Integrase |
| 17 | traE | 18480..19301 [+], 822 | TraE protein | |
| 18 | traF | 19508..20605 [+], 1098 | TraF protein | |
| 19 | ECAGIv1_0021 | 20742..20885 [+], 144 | hypothetical protein | |
| 20 | ECAGIv1_0022 | 21043..21324 [+], 282 | hypothetical protein | |
| 21 | ECAGIv1_0023 | 21945..22184 [+], 240 | hypothetical protein | |
| 22 | dfrA1 | 23681..24154 [+], 474 | dihydrofolate reductase type 1 | AR |
| 23 | sat2 | 24249..24773 [+], 525 | streptothricin acetyltransferase | |
| 24 | aadA1 | 24831..25619 [+], 789 | streptomycin 3''-adenylyltransferase | AR |
| 25 | ECAGIv1_0029 | 25695..26192 [+], 498 | hypothetical protein | |
| 26 | ECAGIv1_0030 | 26253..26624 [+], 372 | hypothetical protein | |
| 27 | ECAGIv1_0031 | 27034..27411 [+], 378 | YbgA protein | |
| 28 | ECAGIv1_0032 | 27461..27586 [+], 126 | putative transposase | |
| 29 | tnsE | 27642..29258 [-], 1617 | transposon Tn7 transposition protein tnsE | |
| 30 | tnsD | 29259..30785 [-], 1527 | transposon Tn7 transposition protein tnsD | |
| 31 | tnsC | 30788..32455 [-], 1668 | transposon Tn7 transposition protein tnsC | |
| 32 | tnsB | 32452..34560 [-], 2109 | transposon Tn7 transposition protein tnsB | |
| 33 | tnsA | 34547..35368 [-], 822 | transposon Tn7 transposition protein tnsA | |
| 34 | ECAGIv1_0040 | 37333..38031 [-], 699 | hypothetical protein | |
| 35 | ECAGIv1_0041 | 38308..39270 [-], 963 | hypothetical protein | |
| 36 | ECAGIv1_0042 | 39378..39812 [-], 435 | hypothetical protein | |
| 37 | ECAGIv1_0044 | 40297..40656 [-], 360 | hypothetical protein | |
| 38 | ECAGIv1_0045 | 40854..41363 [-], 510 | anti-restriction protein | |
| 39 | ECAGIv1_0046 | 41504..41695 [-], 192 | hypothetical protein | |
| 40 | ECAGIv1_0047 | 41782..42168 [-], 387 | hypothetical protein | |
| 41 | ECAGIv1_0048 | 42321..42950 [-], 630 | hypothetical protein | |
| 42 | ECAGIv1_0049 | 43273..44013 [+], 741 | hypothetical protein | |
| 43 | ECAGIv1_0050 | 44017..44934 [-], 918 | putative integrase/DNA breaking-rejoining enzyme | |
| 44 | ECAGIv1_0051 | 46064..46291 [-], 228 | hypothetical protein | |
| 45 | ECAGIv1_0052 | 46461..46736 [+], 276 | hypothetical protein | |
| 46 | ECAGIv1_0053 | 46832..47149 [+], 318 | hypothetical protein | |
| 47 | ECAGIv1_0054 | 47244..47441 [-], 198 | hypothetical protein | |
| 48 | ECAGIv1_0055 | 47926..48189 [-], 264 | putative ribosomal protein | |
| 49 | ECAGIv1_0056 | 48414..48926 [-], 513 | hypothetical protein | |
| 50 | ECAGIv1_0057 | 49284..49490 [+], 207 | hypothetical protein | |
| 51 | ECAGIv1_0058 | 49538..51061 [-], 1524 | putative TraG-like protein | Tfc19, T4SS component |
| 52 | ECAGIv1_0059 | 51058..51417 [-], 360 | putative membrane protein | |
| 53 | ECAGIv1_0060 | 51422..52843 [-], 1422 | putative exported protein | Tfc22, T4SS component |
| 54 | ECAGIv1_0061 | 52850..53824 [-], 975 | putative exported protein | Tfc23, T4SS component |
| 55 | ECAGIv1_0062 | 53821..54216 [-], 396 | hypothetical protein | Tfc24, T4SS component |
| 56 | ECAGIv1_0063 | 54751..56838 [-], 2088 | Iha adhesin | |
| 57 | ECAGIv1_0064 | 56994..57380 [-], 387 | hypothetical protein | Tfc17, T4SS component |
| 58 | ECAGIv1_0065 | 57377..60226 [-], 2850 | putative TraC plasmid-related ATPase | Tfc16, T4SS component |
| 59 | ECAGIv1_0066 | 60226..60630 [-], 405 | putative lipoprotein | Tfc15, T4SS component |
| 60 | ECAGIv1_0067 | 60648..62126 [-], 1479 | putative bacterial conjugation TrbI-like protein | Tfc14, T4SS component |
| 61 | ECAGIv1_0068 | 62116..63036 [-], 921 | putative exported protein | Tfc13, T4SS component |
| 62 | ECAGIv1_0069 | 63033..63680 [-], 648 | hypothetical protein | Tfc12, T4SS component |
| 63 | ECAGIv1_0070 | 63677..64042 [-], 366 | hypothetical protein | Tfc11, T4SS component |
| 64 | ECAGIv1_0071 | 64062..64427 [-], 366 | putative membrane protein | Tfc10, T4SS component |
| 65 | ECAGIv1_0072 | 64455..64697 [-], 243 | hypothetical protein | |
| 66 | ECAGIv1_0073 | 64697..65038 [-], 342 | putative exported protein with RAQPRD domain | Tfc9, T4SS component |
| 67 | ECAGIv1_0074 | 65282..65473 [+], 192 | hypothetical protein | |
| 68 | ECAGIv1_0075 | 65557..66318 [-], 762 | putative transmembrane protein | Tfc8, T4SS component |
| 69 | ECAGIv1_0076 | 66299..68452 [-], 2154 | putative TraD plasmid transfer protein | Tfc6, T4SS component |
| 70 | ECAGIv1_0077 | 68416..69201 [-], 786 | hypothetical protein | |
| 71 | ECAGIv1_0078 | 69198..69632 [-], 435 | hypothetical protein | |
| 72 | ECAGIv1_0079 | 69707..70297 [-], 591 | putative restriction endonuclease | |
| 73 | ECAGIv1_0080 | 70297..70824 [-], 528 | hypothetical protein | Tfc5, T4SS component |
| 74 | ECAGIv1_0081 | 70821..71456 [-], 636 | putative Lytic transglycosylase, catalytic precursor | Tfc4, T4SS component |
| 75 | ECAGIv1_0082 | 71456..72175 [-], 720 | putative exported protein | Tfc3, T4SS component |
| 76 | ECAGIv1_0083 | 72180..72959 [-], 780 | hypothetical protein | Tfc2, T4SS component |
| 77 | ECAGIv1_0084 | 72956..73492 [-], 537 | putative PilL protein | Tfc2, T4SS component |
| 78 | ECAGIv1_0085 | 73725..73910 [-], 186 | hypothetical protein | |
| 79 | ECAGIv1_0086 | 74107..75156 [-], 1050 | putative transposase, YhgA-like protein | |
| 80 | ssb | 75122..75679 [-], 558 | single-stranded DNA-binding protein | |
| 81 | ECAGIv1_0088 | 75692..75895 [-], 204 | hypothetical protein | |
| 82 | ECAGIv1_0089 | 75995..76459 [-], 465 | hypothetical protein | |
| 83 | ECAGIv1_0090 | 77150..79168 [-], 2019 | DNA topoisomerase 3 | |
| 84 | ECAGIv1_0091 | 79185..79910 [-], 726 | hypothetical protein | |
| 85 | ECAGIv1_0092 | 80012..80662 [-], 651 | hypothetical protein | |
| 86 | ECAGIv1_0093 | 80662..81927 [-], 1266 | hypothetical protein | |
| 87 | ECAGIv1_0094 | 82188..82409 [-], 222 | putative Sb34 protein | |
| 88 | ECAGIv1_0095 | 82406..82621 [-], 216 | hypothetical protein | |
| 89 | ECAGIv1_0096 | 82670..83083 [-], 414 | hypothetical protein | |
| 90 | ECAGIv1_0097 | 83094..83333 [-], 240 | hypothetical protein | |
| 91 | ECAGIv1_0098 | 83293..83514 [-], 222 | hypothetical protein | |
| 92 | ECAGIv1_0099 | 83516..83914 [-], 399 | hypothetical protein | |
| 93 | ECAGIv1_0100 | 83911..84426 [-], 516 | hypothetical protein | |
| 94 | ECAGIv1_0101 | 84428..84946 [-], 519 | putative ea22-like protein | |
| 95 | ECAGIv1_0102 | 85014..85235 [-], 222 | putative transcriptional regulator | |
| 96 | ECAGIv1_0103 | 85232..85444 [-], 213 | putative ea22-like protein | |
| 97 | ECAGIv1_0104 | 85441..85617 [-], 177 | hypothetical protein | |
| 98 | ECAGIv1_0105 | 85624..85872 [-], 249 | hypothetical protein | |
| 99 | ECAGIv1_0106 | 85869..86447 [-], 579 | hypothetical protein | |
| 100 | ECAGIv1_0107 | 86444..87175 [-], 732 | hypothetical protein | |
| 101 | ECAGIv1_0108 | 87168..88817 [-], 1650 | hypothetical protein | |
| 102 | dnaB | 88807..90195 [-], 1389 | replicative DNA helicase | |
| 103 | ECAGIv1_0110 | 90329..90880 [+], 552 | hypothetical protein | |
| 104 | ECAGIv1_0111 | 91270..92148 [-], 879 | putative chromosome partitioning protein | |