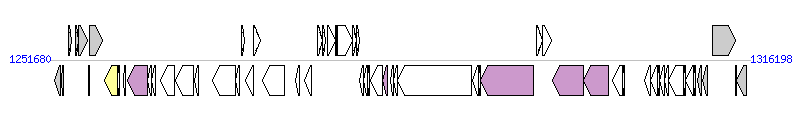The graph information of ICE_Sag2603_rplL components from AE009948 |
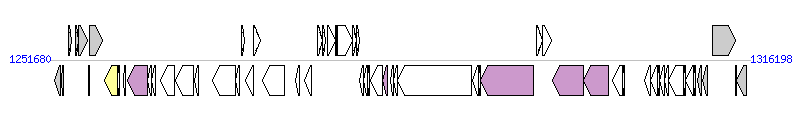 |
| Complete gene list of ICE_Sag2603_rplL from AE009948 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | SAG1238 | 1252057..1252665 [-], 609 | hypothetical protein | |
| 2 | SAG1239 | 1252715..1252945 [-], 231 | conserved hypothetical protein | |
| 3 | SAG1241 | 1253409..1253684 [+], 276 | transposase OrfA, IS3 family | |
| 4 | SAG1243 | 1254003..1254293 [+], 291 | ISSdy1, transposase OrfA | |
| 5 | SAG1244 | 1254329..1255108 [+], 780 | ISSdy1, transposase OrfB | |
| 6 | SAG1245 | 1255213..1255329 [-], 117 | hypothetical protein | |
| 7 | SAG1246 | 1255316..1256485 [+], 1170 | hypothetical protein | |
| 8 | SAG1247 | 1256680..1257879 [-], 1200 | site-specific recombinase, phage integrase family | Integrase |
| 9 | SAG1248 | 1257902..1258129 [-], 228 | conserved hypothetical protein | |
| 10 | SAG1249 | 1258408..1258632 [-], 225 | transcriptional regulator, Cro/CI family | |
| 11 | SAG1250 | 1258813..1260678 [-], 1866 | Tn5252, relaxase | |
| 12 | SAG1251 | 1260665..1261030 [-], 366 | Tn5252, Orf 9 protein | |
| 13 | SAG1252 | 1261037..1261399 [-], 363 | Tn5252, Orf 10 protein | |
| 14 | SAG1253 | 1261820..1263127 [-], 1308 | transposase, ISL3 family | |
| 15 | merA-1 | 1263287..1264927 [-], 1641 | mercuric reductase | |
| 16 | merR-1 | 1264956..1265348 [-], 393 | mercuric resistance operon regulatory protein MerR | |
| 17 | SAG1257 | 1266612..1268741 [-], 2130 | cation-transporting ATPase, E1-E2 family | |
| 18 | cadC | 1268734..1269102 [-], 369 | cadmium efflux system accessory protein | |
| 19 | SAG1259 | 1269305..1269604 [+], 300 | conserved hypothetical protein | |
| 20 | SAG1260 | 1269658..1270446 [-], 789 | hypothetical protein | |
| 21 | SAG1261 | 1270455..1271051 [+], 597 | conserved hypothetical protein | |
| 22 | SAG1262 | 1271225..1273312 [-], 2088 | cation-transporting ATPase, E1-E2 family | |
| 23 | SAG1264 | 1274268..1274714 [-], 447 | transcriptional repressor CopY, putative | |
| 24 | SAG1265 | 1275139..1275759 [-], 621 | cadmium resistance transporter, putative | |
| 25 | SAG1266 | 1276299..1276757 [+], 459 | hypothetical protein | |
| 26 | SAG1267 | 1276791..1277117 [+], 327 | hypothetical protein | |
| 27 | SAG1268 | 1277301..1277993 [+], 693 | repressor protein, putative | |
| 28 | SAG1269 | 1277996..1278130 [+], 135 | hypothetical protein | |
| 29 | SAG1270 | 1278136..1279551 [+], 1416 | ImpB/MucB/SamB family protein | |
| 30 | SAG1271 | 1279548..1279898 [+], 351 | conserved hypothetical protein | |
| 31 | SAG1272 | 1279885..1280193 [+], 309 | conserved hypothetical protein | |
| 32 | SAG1273 | 1280202..1280558 [-], 357 | conserved hypothetical protein | |
| 33 | SAG1274 | 1280548..1280937 [-], 390 | conserved hypothetical protein | |
| 34 | SAG1275 | 1280934..1281161 [-], 228 | hypothetical protein | |
| 35 | SAG1276 | 1281215..1282291 [-], 1077 | conserved hypothetical protein | |
| 36 | SAG1277 | 1282330..1282821 [-], 492 | hypothetical protein | PrgL, T4SS component |
| 37 | SAG1278 | 1283064..1283354 [-], 291 | hypothetical protein | |
| 38 | SAG1279 | 1283368..1283667 [-], 300 | conserved domain protein | |
| 39 | SAG1280 | 1283738..1290562 [-], 6825 | SNF2 family protein | |
| 40 | SAG1281 | 1290611..1291162 [-], 552 | hypothetical protein | |
| 41 | SAG1282 | 1291146..1291337 [-], 192 | calcium-binding protein, putative | |
| 42 | ssp-5 | 1291338..1296233 [-], 4896 | agglutinin receptor | PrgB, T4SS component |
| 43 | abiGI | 1296512..1297102 [+], 591 | abortive infection protein AbiGI | |
| 44 | abiGII | 1297099..1297944 [+], 846 | abortive infection protein AbiGII | |
| 45 | SAG1286 | 1298030..1300831 [-], 2802 | Tn5252, Orf28 | PrgK, T4SS component |
| 46 | SAG1287 | 1300833..1303163 [-], 2331 | Tn5252, Orf26 | PrgJ, T4SS component |
| 47 | SAG1289 | 1303556..1304410 [-], 855 | Tn5252, Orf23 | |
| 48 | SAG1290 | 1304429..1304671 [-], 243 | hypothetical protein | |
| 49 | SAG1292 | 1306506..1306994 [-], 489 | hypothetical protein | |
| 50 | SAG1293 | 1307071..1307655 [-], 585 | protease, putative | |
| 51 | SAG1294 | 1307658..1307891 [-], 234 | conserved hypothetical protein | |
| 52 | SAG1295 | 1307900..1308283 [-], 384 | conserved hypothetical protein | |
| 53 | SAG1296 | 1308294..1308722 [-], 429 | conserved hypothetical protein | |
| 54 | SAG1297 | 1308706..1310061 [-], 1356 | C-5 cytosine-specific DNA methylase | |
| 55 | SAG1298 | 1310113..1310208 [-], 96 | hypothetical protein | |
| 56 | SAG1299 | 1310210..1311028 [-], 819 | conserved hypothetical protein | |
| 57 | SAG1300 | 1311025..1311198 [-], 174 | conserved hypothetical protein | |
| 58 | rplL | 1311397..1311762 [-], 366 | ribosomal protein L7/L12 | |
| 59 | rplJ | 1311826..1312326 [-], 501 | ribosomal protein L10 | |
| 60 | SAG1303 | 1312759..1314867 [+], 2109 | ATP-dependent Clp protease, ATP-binding subunit | |
| 61 | SAG1304 | 1314851..1314949 [-], 99 | hypothetical protein | |
| 62 | SAG1305 | 1314960..1315904 [-], 945 | homocysteine S-methyltransferase MmuM, putative | |