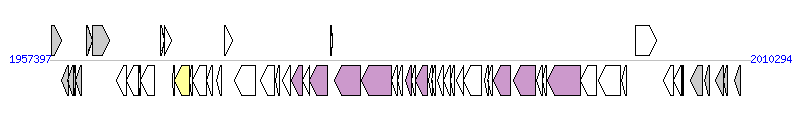The graph information of ICE_2603_tRNALys components from AE009948 |
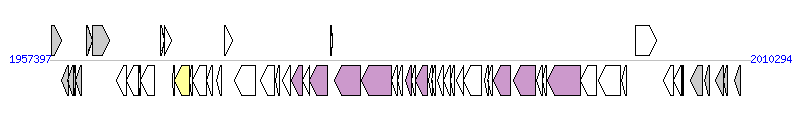 |
| Complete gene list of ICE_2603_tRNALys from AE009948 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | SAG1972 | 1957541..1958257 [+], 717 | transcriptional regulator, MerR family | |
| 2 | SAG1973 | 1958296..1958766 [-], 471 | acetyltransferase, GNAT family | |
| 3 | SAG1974 | 1958738..1959196 [-], 459 | MutT/nudix family protein | |
| 4 | SAG1975 | 1959183..1959326 [-], 144 | hypothetical protein | |
| 5 | SAG1976 | 1959332..1959802 [-], 471 | conserved hypothetical protein | |
| 6 | SAG1977 | 1960129..1960620 [+], 492 | acetyltransferase, GNAT family | |
| 7 | SAG1978 | 1960613..1961881 [+], 1269 | ATPase, AAA family | |
| 8 | SAG1979 | 1962397..1963158 [-], 762 | membrane protein, putative | |
| 9 | SAG1980 | 1963155..1964057 [-], 903 | ABC transporter, ATP-binding protein | |
| 10 | SAG1981 | 1964054..1964260 [-], 207 | hypothetical protein | |
| 11 | SAG1982 | 1964250..1965329 [-], 1080 | transcriptional regulator, Cro/CI family | |
| 12 | SAG1983 | 1965710..1966027 [+], 318 | conserved hypothetical protein | |
| 13 | SAG1984 | 1966024..1966590 [+], 567 | conserved hypothetical protein TIGR00730 | |
| 14 | SAG1985 | 1966644..1966799 [-], 156 | hypothetical protein | |
| 15 | SAG1986 | 1966825..1967952 [-], 1128 | site-specific recombinase, phage integrase family | Integrase |
| 16 | SAG1987 | 1967956..1968141 [-], 186 | conserved hypothetical protein | |
| 17 | SAG1988 | 1968202..1969230 [-], 1029 | conserved hypothetical protein | |
| 18 | SAG1989 | 1969233..1969652 [-], 420 | hypothetical protein | |
| 19 | SAG1990 | 1969978..1970361 [-], 384 | hypothetical protein | |
| 20 | SAG1991 | 1970585..1971199 [+], 615 | transcriptional regulator, Cro/CI family | |
| 21 | SAG1992 | 1971372..1972928 [-], 1557 | protein of unknown function | |
| 22 | SAG1993 | 1973269..1974390 [-], 1122 | site-specific recombinase, phage integrase family | |
| 23 | SAG1994 | 1974440..1974766 [-], 327 | conserved hypothetical protein | |
| 24 | SAG1995 | 1974959..1975591 [-], 633 | hypothetical protein | |
| 25 | SAG1996 | 1975672..1976463 [-], 792 | cell wall surface anchor family protein, putative | PrgC, T4SS component |
| 26 | SAG1997 | 1976480..1977028 [-], 549 | hypothetical protein | |
| 27 | SAG1998 | 1977028..1978401 [-], 1374 | hypothetical protein | PrgK, T4SS component |
| 28 | SAG1999 | 1978564..1978707 [+], 144 | hypothetical protein | |
| 29 | SAG2000 | 1978897..1980897 [-], 2001 | membrane protein, putative | YddG, T4SS component |
| 30 | SAG2001 | 1980913..1983183 [-], 2271 | conjugal transfer protein, interruption-C | YddE, T4SS component |
| 31 | SAG2002 | 1983230..1983619 [-], 390 | IS1381, transposase OrfB | |
| 32 | SAG2003 | 1983652..1984035 [-], 384 | IS1381, transposase OrfA | |
| 33 | SAG2005 | 1984296..1984706 [-], 411 | conserved hypothetical protein | YddD, T4SS component |
| 34 | SAG2006 | 1984717..1984983 [-], 267 | conserved hypothetical protein | |
| 35 | SAG2007 | 1985004..1985957 [-], 954 | conserved hypothetical protein | YddB, T4SS component |
| 36 | SAG2008 | 1986010..1986264 [-], 255 | conserved hypothetical protein | |
| 37 | SAG2009 | 1986257..1986523 [-], 267 | conserved hypothetical protein | |
| 38 | SAG2010 | 1986668..1987141 [-], 474 | hypothetical protein | |
| 39 | SAG2011 | 1987149..1987631 [-], 483 | conserved hypothetical protein | |
| 40 | SAG2012 | 1987684..1987956 [-], 273 | hypothetical protein | |
| 41 | SAG2013 | 1988091..1988660 [-], 570 | hypothetical protein | |
| 42 | SAG2014 | 1988663..1990012 [-], 1350 | hypothetical protein | |
| 43 | SAG2015 | 1990209..1990508 [-], 300 | transcriptional regulator, Cro/CI family | |
| 44 | SAG2016 | 1990492..1990869 [-], 378 | hypothetical protein | |
| 45 | SAG2017 | 1990885..1992174 [-], 1290 | transcriptional regulator, Cro/CI family | Relaxase, MOBT Family |
| 46 | SAG2018 | 1992405..1994066 [-], 1662 | FtsK/SpoIIIE family protein | YdcQ, T4SS component |
| 47 | SAG2019 | 1994143..1994604 [-], 462 | hypothetical protein | |
| 48 | SAG2020 | 1994626..1994922 [-], 297 | hypothetical protein | |
| 49 | SAG2021 | 1994982..1997462 [-], 2481 | cell wall surface anchor family protein | PrgB, T4SS component |
| 50 | SAG2022 | 1997467..1998720 [-], 1254 | transposase, ISL3 family | |
| 51 | merA-2 | 1998880..2000520 [-], 1641 | mercuric reductase | |
| 52 | merR-2 | 2000549..2000941 [-], 393 | mercuric resistance operon regulatory protein MerR | |
| 53 | SAG2025 | 2001647..2003215 [+], 1569 | Mn2+/Fe2+ transporter, NRAMP family | |
| 54 | SAG2026 | 2003783..2004505 [-], 723 | membrane protein, putative | |
| 55 | SAG2027 | 2004509..2005126 [-], 618 | ABC transporter, ATP-binding protein | |
| 56 | SAG2028 | 2005184..2005294 [-], 111 | conserved hypothetical protein | |
| 57 | SAG2029 | 2005813..2006667 [-], 855 | streptomycin resistance protein | |
| 58 | SAG2030 | 2006846..2007238 [-], 393 | hypothetical protein | |
| 59 | SAG2031 | 2007673..2008281 [-], 609 | hypothetical protein | |
| 60 | SAG2032 | 2008268..2008603 [-], 336 | conserved hypothetical protein | |
| 61 | SAG2033 | 2009125..2009613 [-], 489 | acetyltransferase, GNAT family | |