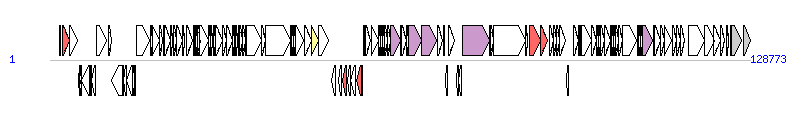The graph information of CMGEYY060816 components from KX077898 |
 |
| Complete gene list of CMGEYY060816 from KX077898 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 1746..1916 [+], 171 | hypothetical protein | |
| 2 | mefA | 2303..3520 [+], 1218 | macrolide-efflux protein | AR |
| 3 | msrD | 3640..5103 [+], 1464 | ATPase component of ABC transporter | |
| 4 | - | 5221..5520 [-], 300 | hypothetical protein | |
| 5 | - | 5507..5875 [-], 369 | hypothetical protein | |
| 6 | - | 5872..7287 [-], 1416 | ImpB/MucB/SamB family protein | |
| 7 | - | 7292..7420 [-], 129 | hypothetical protein | |
| 8 | - | 7410..7637 [-], 228 | hypothetical protein | |
| 9 | - | 7640..8329 [-], 690 | pleiotropic regulator of exopolysaccharide | |
| 10 | - | 8605..10428 [+], 1824 | hypothetical protein | |
| 11 | - | 10762..11331 [+], 570 | hypothetical protein | |
| 12 | - | 11370..13325 [-], 1956 | DNA polymerase I | |
| 13 | - | 13374..13541 [-], 168 | hypothetical protein | |
| 14 | - | 13560..14108 [-], 549 | hypothetical protein | |
| 15 | - | 14113..15153 [-], 1041 | hypothetical protein | |
| 16 | - | 15227..15550 [-], 324 | hypothetical protein | |
| 17 | - | 15525..15659 [-], 135 | hypothetical protein | |
| 18 | - | 15949..18234 [+], 2286 | DNA primase/helicase | |
| 19 | - | 18519..18800 [+], 282 | hypothetical protein | |
| 20 | - | 18781..20157 [+], 1377 | DNA helicase | |
| 21 | - | 20150..20626 [+], 477 | hypothetical protein | |
| 22 | - | 20821..21003 [+], 183 | hypothetical protein | |
| 23 | - | 21048..22085 [+], 1038 | S-adenosylmethionine synthetase | |
| 24 | - | 22087..22452 [+], 366 | holin | |
| 25 | - | 22751..23212 [+], 462 | zinc-finger protein | |
| 26 | - | 23184..24437 [+], 1254 | adenine-specific methyltransferase | |
| 27 | - | 24437..24670 [+], 234 | hypothetical protein | |
| 28 | - | 25023..26297 [+], 1275 | DNA-cytosine methyltransferase | |
| 29 | - | 26343..26624 [+], 282 | hypothetical protein | |
| 30 | - | 26626..26838 [+], 213 | hypothetical protein | |
| 31 | - | 26840..27070 [+], 231 | hypothetical protein | |
| 32 | - | 27122..27595 [+], 474 | terminase small subunit | |
| 33 | - | 27592..29184 [+], 1593 | terminase large subunit | |
| 34 | - | 29244..29546 [+], 303 | plasmid stabilization system antitoxin protein | |
| 35 | - | 29546..29884 [+], 339 | death on curing protein | |
| 36 | - | 30021..30365 [+], 345 | hypothetical protein | |
| 37 | - | 30423..31706 [+], 1284 | portal protein | |
| 38 | - | 31699..32394 [+], 696 | protease-like protein | |
| 39 | - | 32413..33618 [+], 1206 | major capsid protein | |
| 40 | - | 33618..33878 [+], 261 | hypothetical protein | |
| 41 | - | 33878..34216 [+], 339 | hypothetical protein | |
| 42 | - | 34209..34577 [+], 369 | hypothetical protein | |
| 43 | - | 34599..34910 [+], 312 | hypothetical protein | |
| 44 | - | 34912..35481 [+], 570 | hypothetical protein | |
| 45 | - | 35492..35911 [+], 420 | hypothetical protein | |
| 46 | - | 35944..36075 [+], 132 | hypothetical protein | |
| 47 | - | 36094..38952 [+], 2859 | tail length tape-measure protein | |
| 48 | - | 38949..39671 [+], 723 | hypothetical protein | |
| 49 | - | 39673..44220 [+], 4548 | endopeptidase | |
| 50 | - | 44233..44439 [+], 207 | hypothetical protein | |
| 51 | - | 44445..44813 [+], 369 | hypothetical protein | |
| 52 | - | 44791..45186 [+], 396 | holin | |
| 53 | - | 45193..46662 [+], 1470 | cell wall hydrolase | |
| 54 | - | 46885..48126 [+], 1242 | recombinase | |
| 55 | - | 48089..49381 [+], 1293 | site-specific recombinase | Integrase |
| 56 | - | 49461..51197 [+], 1737 | hypothetical protein | |
| 57 | - | 51839..52498 [-], 660 | 5-amino-6-(5-phosphoribosylamino)uracil reductase | |
| 58 | - | 52989..53561 [-], 573 | putative transcriptional regulator | |
| 59 | - | 53837..54631 [-], 795 | aminoglycoside phosphotransferase | AR |
| 60 | - | 54724..55266 [-], 543 | streptothricin acetyltransferase | |
| 61 | - | 55263..56171 [-], 909 | putative aminoglycoside 6-adenylyltansferase | |
| 62 | - | 56476..57213 [-], 738 | rRNA (adenine-N(6)-) -methyltransferase | AR |
| 63 | - | 57387..57506 [-], 120 | hypothetical protein | |
| 64 | - | 57691..57858 [+], 168 | hypothetical protein | |
| 65 | - | 57935..58102 [+], 168 | hypothetical protein | |
| 66 | - | 58105..58923 [+], 819 | hypothetical protein | |
| 67 | - | 59120..60427 [+], 1308 | DNA-cytosine methyltransferase | |
| 68 | - | 60411..60839 [+], 429 | hypothetical protein | |
| 69 | - | 60850..61233 [+], 384 | hypothetical protein | |
| 70 | - | 61242..61475 [+], 234 | hypothetical protein | |
| 71 | - | 61478..62062 [+], 585 | hypothetical protein | |
| 72 | - | 62103..62627 [+], 525 | hypothetical protein | |
| 73 | - | 62627..64444 [+], 1818 | hypothetical protein | T4CP |
| 74 | - | 64462..64704 [+], 243 | hypothetical protein | |
| 75 | - | 64723..65577 [+], 855 | hypothetical protein | |
| 76 | - | 65639..65992 [+], 354 | hypothetical protein | |
| 77 | - | 65970..68300 [+], 2331 | TrsE-like protein | PrgJ, T4SS component |
| 78 | - | 68302..71103 [+], 2802 | hypothetical protein | PrgK, T4SS component |
| 79 | - | 71240..72250 [+], 1011 | hypothetical protein | |
| 80 | - | 72360..72647 [+], 288 | hypothetical protein | PrgK, T4SS component |
| 81 | - | 72733..73197 [-], 465 | abortive infection protein AbiGII | |
| 82 | - | 73241..74362 [+], 1122 | mobile element protein | |
| 83 | - | 74736..75185 [-], 450 | abortive infection protein AbiGII | |
| 84 | - | 75182..75772 [-], 591 | abortive infection protein AbiGI | |
| 85 | - | 75942..80837 [+], 4896 | agglutinin receptor | PrgB, T4SS component |
| 86 | - | 80838..81029 [+], 192 | putative calcium-binding protein | |
| 87 | - | 81013..81564 [+], 552 | hypothetical protein | |
| 88 | - | 81637..87561 [+], 5925 | SNF2 family protein | |
| 89 | - | 87542..87916 [+], 375 | TnpV | |
| 90 | - | 88283..90202 [+], 1920 | tetracycline resistance protein TetO | AR |
| 91 | - | 90253..91473 [+], 1221 | tetracycline resistance protein | AR |
| 92 | - | 91946..92581 [+], 636 | hypothetical protein | |
| 93 | - | 92676..93101 [+], 426 | hypothetical protein | |
| 94 | - | 93185..93658 [+], 474 | RNA polymerase ECF-type sigma factor | |
| 95 | - | 93811..94842 [+], 1032 | hypothetical protein | |
| 96 | - | 95062..95322 [-], 261 | hypothetical protein | |
| 97 | - | 96334..97188 [+], 855 | chromosome (plasmid) partitioning protein ParB | |
| 98 | - | 97265..97495 [+], 231 | bacterial seryl-tRNA synthetase related | |
| 99 | - | 97566..97757 [+], 192 | hypothetical protein | |
| 100 | - | 97811..99583 [+], 1773 | site-specific recombinase | |
| 101 | - | 99598..100509 [+], 912 | SNF2 family protein | |
| 102 | - | 100580..100879 [+], 300 | hypothetical protein | |
| 103 | - | 100893..101183 [+], 291 | hypothetical protein | |
| 104 | - | 101278..101916 [+], 639 | hypothetical protein | |
| 105 | - | 101956..103056 [+], 1101 | hypothetical protein | |
| 106 | - | 103084..103311 [+], 228 | hypothetical protein | |
| 107 | - | 103308..103697 [+], 390 | methyl-accepting chemotaxis protein | |
| 108 | - | 103747..104037 [+], 291 | hypothetical protein | |
| 109 | - | 104107..104583 [+], 477 | putative transcription regulator | |
| 110 | - | 104583..105350 [+], 768 | zeta toxin | |
| 111 | - | 105351..107894 [+], 2544 | hypothetical protein | |
| 112 | - | 108000..108122 [+], 123 | hypothetical protein | |
| 113 | - | 108232..108594 [+], 363 | hypothetical protein | |
| 114 | - | 108597..108962 [+], 366 | hypothetical protein | |
| 115 | - | 108949..110832 [+], 1884 | relaxase | Relaxase, MOBP Family |
| 116 | - | 110929..111144 [+], 216 | hypothetical protein | |
| 117 | - | 111226..112302 [+], 1077 | epidermin leader peptide processing serine | |
| 118 | - | 112343..113041 [+], 699 | two-component response regulator | |
| 119 | - | 113022..114362 [+], 1341 | sensor-receptor histidine kinase NisK | |
| 120 | - | 114546..115247 [+], 702 | hypothetical protein | |
| 121 | - | 115240..115977 [+], 738 | arginine/ornithine antiporter ArcD | |
| 122 | - | 115955..116641 [+], 687 | hypothetical protein | |
| 123 | - | 117390..120362 [+], 2973 | lanthionine biosynthesis protein LanB | |
| 124 | - | 120343..122151 [+], 1809 | subtilin transport ATP-binding protein spaT | |
| 125 | - | 122144..123421 [+], 1278 | lanthionine biosynthesis cyclase LanC | |
| 126 | - | 123423..124139 [+], 717 | hypothetical protein | |
| 127 | - | 124407..124811 [+], 405 | hypothetical protein | |
| 128 | - | 125104..125262 [+], 159 | hypothetical protein | |
| 129 | - | 125316..127247 [+], 1932 | site-specific recombinase | |
| 130 | - | 127661..128773 [+], 1113 | UDP-galactopyranose mutase | |