The graph information of ICEclc(JS705) components from AJ006307 |
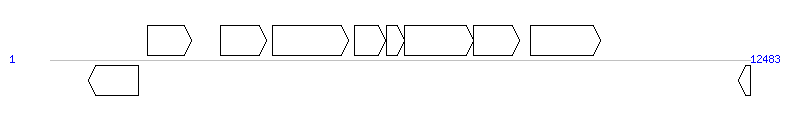 |
| Incomplete gene list of ICEclc(JS705) from AJ006307 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | clcR | 687..1571 [-], 885 | ClcR | |
| 2 | clcA | 1741..2523 [+], 783 | chlorocatechol 1,2-dioxygenase | |
| 3 | mcbF | 3036..3866 [+], 831 | 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase | |
| 4 | mcbAa | 3960..5315 [+], 1356 | terminal oxygenase large subunit | |
| 5 | mcbAb | 5426..5989 [+], 564 | terminal oxygenase small subunit | |
| 6 | mcbAc | 6000..6323 [+], 324 | ferredoxin | |
| 7 | mcbAd | 6323..7555 [+], 1233 | reductase | |
| 8 | mcbB | 7552..8379 [+], 828 | cis-chlorobenzene dihydrodiol dehydrogenase | |
| 9 | tnpA | 8574..9818 [+], 1245 | transposase | |
| 10 | clcR | 12271..12483 [-], 213 | transcriptional activator | |
