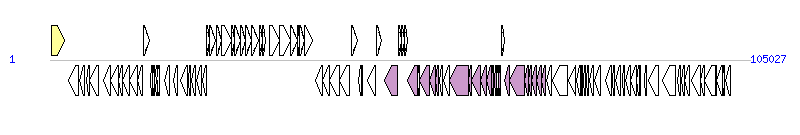The graph information of ICEclc(B13) components from AJ617740 |
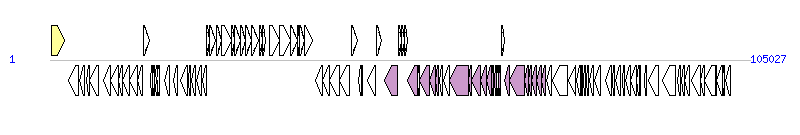 |
| Complete gene list of ICEclc(B13) from AJ617740 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | intB13 | 262..2235 [+], 1974 | integrase | Integrase |
| 2 | - | 2848..4233 [-], 1386 | putative permease | |
| 3 | - | 4438..5157 [-], 720 | oxidoreductase | |
| 4 | - | 5512..5991 [-], 480 | putative ring dioxygenase beta | |
| 5 | - | 5994..7256 [-], 1263 | large subunit aromatic dioxygenase | |
| 6 | - | 8052..9035 [-], 984 | hypothetical protein | |
| 7 | clcE | 9151..10209 [-], 1059 | maleylacetate reductase | |
| 8 | clcD | 10206..10916 [-], 711 | dienelactone hydrolase | |
| 9 | - | 10938..11921 [-], 984 | hypothetical protein | |
| 10 | clcB | 11948..13060 [-], 1113 | chloromuconate cycloisomerase | |
| 11 | clcA | 13057..13839 [-], 783 | chlorocatechol 1,2-dioxygenase | |
| 12 | clcR | 14009..14893 [+], 885 | LysR family transcriptional regulator | |
| 13 | - | 15037..15387 [-], 351 | putative threonine efflux protein | |
| 14 | - | 15405..15647 [-], 243 | hypothetical protein | |
| 15 | - | 15675..16103 [-], 429 | chlorocatechol 1,2-dioxygenase | |
| 16 | - | 16054..16494 [-], 441 | putative outer membrane protein | |
| 17 | - (tciR) | 17162..17959 [-], 798 | transcriptional regulator | TciR; global regulator
for ICEclc activation; [PMID: 24945944] |
| 18 | - (mfsR) | 18502..19188 [-], 687 | transcriptional regulator | MfsR; global regulator
for ICEclc activation; [PMID: 24945944] |
| 19 | - | 19619..20563 [-], 945 | hypothetical protein | |
| 20 | - | 20709..21128 [-], 420 | hypothetical protein | |
| 21 | - | 21241..21900 [-], 660 | TetR-type transcriptional regulator | |
| 22 | - | 21922..22674 [-], 753 | hypothetical protein | |
| 23 | - | 22813..23430 [-], 618 | putative aminophenol repressor | |
| 24 | - | 23526..23939 [+], 414 | ferredoxin-like protein | |
| 25 | - | 23951..24865 [+], 915 | uncharacterized conserved protein | |
| 26 | - | 24910..25722 [+], 813 | 2-aminophenol 1,6-dioxygenase alpha subunit | |
| 27 | - | 25781..27259 [+], 1479 | 2-aminomuconic semialdehyde dehydrogenase | |
| 28 | - | 27249..27701 [+], 453 | 2-aminomuconate deaminase | |
| 29 | - | 27716..28525 [+], 810 | putative 2-keto-4-pentenoate hydratase | |
| 30 | - | 28522..29286 [+], 765 | putative 4-oxalocrotonate decarboxylase | |
| 31 | - | 29347..30288 [+], 942 | acetylating aldehyde dehydrogenase | |
| 32 | - | 30304..31341 [+], 1038 | putative 4-hydroxy-2-ketovalerate aldolase | |
| 33 | - | 31453..31953 [+], 501 | hypothetical protein | |
| 34 | - | 31950..32345 [+], 396 | hypothetical protein | |
| 35 | - | 32963..34498 [+], 1536 | putative outer membrane protein or channel-forming component | |
| 36 | - | 34495..36069 [+], 1575 | putative permease of the major facilitator superfamily | |
| 37 | - | 36077..37111 [+], 1035 | putative multidrug efflux pump | |
| 38 | - | 37143..37445 [+], 303 | hypothetical protein | |
| 39 | - | 37489..38133 [+], 645 | hypothetical protein | |
| 40 | - | 38184..39365 [+], 1182 | putative esterase of the alpha-beta hydrolase superfamily | |
| 41 | - | 39860..40894 [-], 1035 | hypothetical protein | |
| 42 | - | 40922..41917 [-], 996 | predicted amidohydrolase/nitrilase | |
| 43 | - | 41973..43385 [-], 1413 | putative acyl-CoA synthetase | |
| 44 | - | 43387..44967 [-], 1581 | putative acyl-CoA synthetase | |
| 45 | - | 45180..46136 [+], 957 | putative transcriptional regulator | |
| 46 | - | 46315..46698 [-], 384 | hypothetical protein | |
| 47 | - | 46777..46932 [-], 156 | transposase | |
| 48 | - | 47630..48811 [-], 1182 | putative transport protein | |
| 49 | - | 48922..49725 [+], 804 | putative transcriptional regulator | |
| 50 | - | 50240..52087 [-], 1848 | hypothetical protein | Relaxase, MOBH Family |
| 51 | - | 52324..52710 [+], 387 | hypothetical protein | |
| 52 | - | 52710..53168 [+], 459 | hypothetical protein | |
| 53 | - | 53196..53573 [+], 378 | hypothetical protein | |
| 54 | - | 53587..55104 [-], 1518 | hypothetical protein | Tfc19, T4SS component |
| 55 | - | 55120..55479 [-], 360 | hypothetical protein | |
| 56 | - | 55476..56873 [-], 1398 | hypothetical protein | Tfc22, T4SS component |
| 57 | - | 56883..57830 [-], 948 | hypothetical protein | Tfc23, T4SS component |
| 58 | - | 57827..58273 [-], 447 | hypothetical protein | Tfc24, T4SS component |
| 59 | - | 58432..58926 [-], 495 | putative DNA repair protein | |
| 60 | - | 59110..59874 [-], 765 | putative protein-disulfide isomerase | |
| 61 | - | 59888..62755 [-], 2868 | conserved hypothetical protein with VirB4 domain | Tfc16, T4SS component |
| 62 | - | 62755..63195 [-], 441 | hypothetical protein | Tfc15, T4SS component |
| 63 | - | 63176..64594 [-], 1419 | hypothetical protein | Tfc14, T4SS component |
| 64 | - | 64584..65516 [-], 933 | hypothetical protein | Tfc13, T4SS component |
| 65 | - | 65513..66205 [-], 693 | hypothetical protein | Tfc12, T4SS component |
| 66 | - | 66202..66612 [-], 411 | hypothetical protein | Tfc11, T4SS component |
| 67 | - | 66625..66984 [-], 360 | hypothetical protein | Tfc10, T4SS component |
| 68 | - | 67001..67234 [-], 234 | hypothetical protein | |
| 69 | - | 67231..67614 [-], 384 | hypothetical protein | Tfc9, T4SS component |
| 70 | - | 67800..68204 [+], 405 | hypothetical protein | |
| 71 | - | 68241..68990 [-], 750 | hypothetical protein | Tfc8, T4SS component |
| 72 | - | 68987..71173 [-], 2187 | hypothetical protein | Tfc6, T4SS component |
| 73 | - | 71178..71726 [-], 549 | hypothetical protein | Tfc5, T4SS component |
| 74 | - | 71723..72313 [-], 591 | hypothetical protein | Tfc4, T4SS component |
| 75 | - | 72295..73014 [-], 720 | hypothetical protein | Tfc3, T4SS component |
| 76 | - | 73029..73679 [-], 651 | hypothetical protein | Tfc2, T4SS component |
| 77 | - | 73676..74296 [-], 621 | hypothetical protein | Tfc2, T4SS component |
| 78 | - | 74436..75305 [-], 870 | hypothetical protein | |
| 79 | - | 75419..77698 [-], 2280 | putative DNA/RNA helicase | |
| 80 | - | 77798..78907 [-], 1110 | hypothetical protein | |
| 81 | - | 78972..79622 [-], 651 | hypothetical protein | |
| 82 | - | 79699..79959 [-], 261 | hypothetical protein | |
| 83 | - | 79976..80383 [-], 408 | hypothetical protein | |
| 84 | - | 80480..80812 [-], 333 | plasmid protein | |
| 85 | - | 80908..81597 [-], 690 | hypothetical protein | |
| 86 | - | 81655..82572 [-], 918 | hypothetical protein | |
| 87 | - | 83350..84192 [-], 843 | phage-related protein | |
| 88 | - | 84338..84691 [-], 354 | hypothetical protein | |
| 89 | - | 84835..85647 [-], 813 | hypothetical protein | |
| 90 | - | 85934..86212 [-], 279 | hypothetical protein | |
| 91 | - | 86310..87047 [-], 738 | hypothetical protein | |
| 92 | - | 87127..87939 [-], 813 | hypothetical protein | |
| 93 | - | 87986..88378 [-], 393 | hypothetical protein | |
| 94 | - | 88400..88612 [-], 213 | hypothetical protein | |
| 95 | - | 89247..89501 [-], 255 | hypothetical protein | |
| 96 | - | 89746..91347 [-], 1602 | putative DNA methyltransferase | |
| 97 | - | 91884..93896 [-], 2013 | putative DNA topoisomerase III | |
| 98 | - (ssb) | 94175..94615 [-], 441 | putative single-stranded DNA binding protein | Ssb; single-strand DNA binding protein; [PMID: 19943900] |
| 99 | inrR | 94689..95216 [-], 528 | probable transcriptional regulator | |
| 100 | - | 95213..95992 [-], 780 | hypothetical protein | |
| 101 | - | 96323..97567 [-], 1245 | hypothetical protein | |
| 102 | - | 97571..98131 [-], 561 | hypothetical protein | |
| 103 | - (parB) | 98147..99799 [-], 1653 | hypothetical protein | ParB; bacterial partitioning protein; [PMID: 11522360] |
| 104 | - (shi) | 99792..100049 [-], 258 | hypothetical protein | Shi; Impairment of cell division; [PMID: 23333318] |
| 105 | - (parA) | 100033..100908 [-], 876 | hypothetical protein | ParA; bacterial partitioning protein; [PMID: 11522360] |
| 106 | - (alpA) | 100952..101164 [-], 213 | putative transcriptional regulator | AlpA; outer membrane (OM) protein; [PMID: 10200971] |
| 107 | - | 101284..102039 [-], 756 | hypothetical protein | |