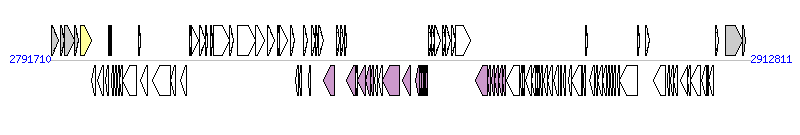The graph information of ICEPaeLESB58-1 components from FM209186 |
 |
| Complete gene list of ICEPaeLESB58-1 from FM209186 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | pqsH | 2792010..2793158 [+], 1149 | probable FAD-dependent monooxygenase | |
| 2 | gacA | 2793500..2794144 [+], 645 | response regulator GacA | |
| 3 | uvrC | 2794145..2795971 [+], 1827 | excinuclease ABC subunit C | |
| 4 | pgsA | 2796005..2796565 [+], 561 | CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase | |
| 5 | PLES_26051 | 2796936..2798879 [+], 1944 | phage-related integrase | Integrase |
| 6 | PLES_26061 | 2798946..2799587 [-], 642 | transcriptional regulator, LysR family | |
| 7 | PLES_26071 | 2800012..2800905 [-], 894 | transcriptional regulator, LysR family | |
| 8 | PLES_26081 | 2801226..2801822 [-], 597 | 4-hydroxyphenylpyruvate dioxygenase (pseudogene) | |
| 9 | PLES_26091 | 2801863..2802039 [+], 177 | transcriptional regulator, AsnC family (pseudogene) | |
| 10 | PLES_26101 | 2802140..2802334 [+], 195 | Heavy metal transport/detoxification protein | |
| 11 | PLES_26111 | 2802409..2802636 [-], 228 | glutaredoxin (pseudogene) | |
| 12 | PLES_26121 | 2802873..2803145 [-], 273 | glutaredoxin | |
| 13 | fdx | 2803369..2803707 [-], 339 | [2Fe-2S] ferredoxin | |
| 14 | PLES_26141 | 2803723..2804202 [-], 480 | transcriptional regulator, MerR family | |
| 15 | PLES_26151 | 2804218..2806647 [-], 2430 | hypothetical protein | |
| 16 | PLES_26161 | 2807081..2807305 [+], 225 | hypothetical protein | |
| 17 | PLES_26171 | 2807415..2808572 [-], 1158 | dihydrolipoamide dehydrogenase | |
| 18 | PLES_26181 | 2809400..2812546 [-], 3147 | heavy metal efflux pump, CzcA family | |
| 19 | PLES_26191 | 2812560..2813459 [-], 900 | hypothetical protein | |
| 20 | PLES_26201 | 2814258..2815349 [-], 1092 | outer membrane efflux protein (pseudogene) | |
| 21 | PLES_26211 | 2815869..2816126 [+], 258 | protein of unknown function DUF156 | |
| 22 | PLES_26221 | 2816136..2817446 [+], 1311 | putative nrbE-like protein | |
| 23 | efflux | 2817640..2818602 [+], 963 | cation efflux family protein | |
| 24 | PLES_26241 | 2818646..2819023 [+], 378 | transcriptional regulator, ArsR family | |
| 25 | PLES_26251 | 2819556..2819822 [+], 267 | hypothetical protein | |
| 26 | PLES_26261 | 2819989..2822703 [+], 2715 | heavy metal translocating P-type ATPase | |
| 27 | PLES_26271 | 2822815..2823417 [+], 603 | efflux transporter, RND family, MFP subunit | |
| 28 | PLES_26281 | 2824136..2827312 [+], 3177 | transporter, hydrophobe/amphiphile efflux-1 (HAE1) family | |
| 29 | PLES_26291 | 2827333..2828838 [+], 1506 | RND efflux system, outer membrane lipoprotein, NodT family | |
| 30 | PLES_26301 | 2829354..2830535 [+], 1182 | major facilitator superfamily MFS_1 | |
| 31 | PLES_26311 | 2830987..2831127 [+], 141 | hypothetical protein | |
| 32 | PLES_26321 | 2831394..2832848 [+], 1455 | mercuric reductase | |
| 33 | PLES_26331 | 2833295..2834071 [+], 777 | major facilitator superfamily MFS_1 (pseudogene) | |
| 34 | PLES_26341 | 2834118..2834621 [-], 504 | hypothetical protein | |
| 35 | PLES_26351 | 2834803..2835192 [-], 390 | hypothetical protein | |
| 36 | PLES_26361 | 2835545..2836240 [+], 696 | transcriptional regulator, GntR family | |
| 37 | PLES_26371 | 2836396..2836806 [-], 411 | transcriptional regulator, GntR family | |
| 38 | PLES_26381 | 2836987..2837688 [+], 702 | putative exported protein | |
| 39 | PLES_26391 | 2837699..2837956 [+], 258 | glutaredoxin | |
| 40 | PLES_26401 | 2838131..2839003 [+], 873 | 2-dehydro-3-deoxyphosphooctonate aldolase | |
| 41 | PLES_26421 | 2839054..2840925 [-], 1872 | Relaxase | Relaxase, MOBH Family |
| 42 | PLES_26431 | 2841242..2841868 [+], 627 | hypothetical protein | |
| 43 | PLES_26441 | 2841881..2842513 [+], 633 | hypothetical protein | |
| 44 | PLES_26451 | 2842598..2842963 [+], 366 | conserved hypothetical protein | |
| 45 | PLES_26461 | 2842975..2844495 [-], 1521 | STY4579 | Tfc19, T4SS component |
| 46 | PLES_26471 | 2844511..2844882 [-], 372 | hypothetical protein | |
| 47 | PLES_26481 | 2844879..2846276 [-], 1398 | conserved hpothetical protein | Tfc22, T4SS component |
| 48 | PLES_26491 | 2846286..2847233 [-], 948 | conserved hypothetical protein | Tfc23, T4SS component |
| 49 | PLES_26501 | 2847230..2847622 [-], 393 | hypothetical protein | Tfc24, T4SS component |
| 50 | PLES_26511 | 2847839..2848333 [-], 495 | RadC family DNA repair protein | |
| 51 | PLES_26521 | 2848516..2849301 [-], 786 | conserved hypothetical protein | |
| 52 | PLES_26531 | 2849315..2852200 [-], 2886 | conserved hypothetical protein | Tfc16, T4SS component |
| 53 | PLES_26541 | 2852621..2854039 [-], 1419 | conserved hypothetical protein | Tfc14, T4SS component |
| 54 | PLES_26551 | 2854937..2855629 [-], 693 | conserved hypothetical protein | Tfc12, T4SS component |
| 55 | PLES_26561 | 2855626..2855958 [-], 333 | hypothetical protein | Tfc11, T4SS component |
| 56 | PLES_26571 | 2856049..2856408 [-], 360 | conserved hypothetical protein | Tfc10, T4SS component |
| 57 | PLES_26581 | 2856425..2856658 [-], 234 | hypothetical protein | |
| 58 | PLES_26591 | 2856655..2857038 [-], 384 | plasmid Ignore, RAQPRD family | Tfc9, T4SS component |
| 59 | PLES_26601 | 2857242..2857712 [+], 471 | DNA binding domain, excisionase family | |
| 60 | PLES_26611 | 2857709..2858284 [+], 576 | conserved hypothetical protein | |
| 61 | PLES_26621 | 2858302..2859216 [+], 915 | AAA ATPase, central domain protein | |
| 62 | PLES_26631 | 2859680..2860180 [+], 501 | hypothetical protein | |
| 63 | PLES_26641 | 2860180..2861082 [+], 903 | hypothetical protein | |
| 64 | PLES_26651 | 2861121..2861846 [+], 726 | hypothetical protein | |
| 65 | PLES_26661 | 2861898..2864480 [+], 2583 | COG1196: Chromosome segregation ATPases | |
| 66 | PLES_26671 | 2865267..2867456 [-], 2190 | conserved hypothetical protein | Tfc6, T4SS component |
| 67 | PLES_26681 | 2867461..2867964 [-], 504 | conserved hypothetical protein | Tfc5, T4SS component |
| 68 | PLES_26691 | 2868006..2868575 [-], 570 | conserved hypothetical protein | Tfc4, T4SS component |
| 69 | PLES_26701 | 2868578..2869315 [-], 738 | conserved hypothetical protein | Tfc3, T4SS component |
| 70 | PLES_26711 | 2869328..2869972 [-], 645 | putative secreted protein | Tfc2, T4SS component |
| 71 | PLES_26721 | 2869969..2870430 [-], 462 | putative secreted protein | Tfc2, T4SS component |
| 72 | PLES_26731 | 2870707..2872986 [-], 2280 | conserved hypothetical protein | |
| 73 | PLES_26741 | 2873123..2873428 [-], 306 | hypothetical protein | |
| 74 | PLES_26751 | 2873518..2873838 [-], 321 | hypothetical protein | |
| 75 | PLES_26761 | 2873889..2874998 [-], 1110 | conserved hypothetical, plasmid-related protein | |
| 76 | PLES_26771 | 2875063..2875710 [-], 648 | hypothetical protein | |
| 77 | PLES_26781 | 2875787..2876047 [-], 261 | conserved Hypothetical protein | |
| 78 | PLES_26791 | 2876064..2876471 [-], 408 | conserved Hypothetical protein | |
| 79 | PLES_26801 | 2876576..2876917 [-], 342 | conserved plasmid protein | |
| 80 | PLES_26811 | 2877012..2877701 [-], 690 | hypothetical protein | |
| 81 | PLES_26821 | 2877796..2878623 [-], 828 | protein of unknown function DUF932 | |
| 82 | PLES_26831 | 2878769..2879767 [-], 999 | hypothetical protein | |
| 83 | PLES_26841 | 2879985..2880269 [-], 285 | hypothetical protein | |
| 84 | PLES_26851 | 2880577..2880837 [-], 261 | hypothetical protein | |
| 85 | PLES_26861 | 2881389..2881904 [-], 516 | Tn3 family transposase (pseudogene) | |
| 86 | merA | 2881923..2883611 [-], 1689 | Mercuric reductase MerA | |
| 87 | merP | 2883622..2883909 [-], 288 | Periplasmic mercuric ion binding protein, MerP | |
| 88 | merT | 2883922..2884272 [-], 351 | Mercuric transport protein MerT | |
| 89 | merR | 2884344..2884751 [+], 408 | Regulatory protein merR | |
| 90 | PLES_26911 | 2885013..2885837 [-], 825 | COG1100: GTPase SAR1 and related small G proteins | |
| 91 | PLES_26921 | 2886126..2886404 [-], 279 | COG0528: Uridylate kinase | |
| 92 | PLES_26931 | 2886502..2887239 [-], 738 | COG0834: ABC-type amino acid transport/signal transduction systems, periplasmic component/domain | |
| 93 | ORF C92 | 2887323..2888009 [-], 687 | hypothetical protein | |
| 94 | PLES_26951 | 2888192..2888584 [-], 393 | hypothetical protein | |
| 95 | PLES_26961 | 2888606..2889019 [-], 414 | conserved hypothetical protein | |
| 96 | PLES_26971 | 2889156..2889467 [-], 312 | hypothetical protein | |
| 97 | PLES_26981 | 2889804..2890304 [-], 501 | putative lipoprotein signal peptidase LspA | |
| 98 | PLES_26991 | 2890308..2893262 [-], 2955 | Heavy metal translocating P-type ATPase | |
| 99 | metR | 2893315..2893710 [+], 396 | transcriptional regulator, MerR family | |
| 100 | PLES_27002 | 2894786..2895421 [+], 636 | Co/Zn/Cd efflux system component | |
| 101 | PLES_27011 | 2896145..2898175 [-], 2031 | DNA topoisomerase III | |
| 102 | PLES_27021 | 2898459..2898899 [-], 441 | Putative single-stranded DNA binding protein | |
| 103 | PLES_27031 | 2898973..2899500 [-], 528 | Putative integrase regulator R protein | |
| 104 | PLES_27041 | 2899497..2900288 [-], 792 | Domain of unknown function DUF1845 | |
| 105 | PLES_27051 | 2900718..2901956 [-], 1239 | hypothetical protein | |
| 106 | PLES_27061 | 2901960..2902520 [-], 561 | COG0635: Coproporphyrinogen III oxidase and related Fe-S oxidoreductases | |
| 107 | PLES_27071 | 2902535..2904214 [-], 1680 | COG1475: Predicted transcriptional regulators | |
| 108 | PLES_27081 | 2904460..2905335 [-], 876 | Cobyrinic acid a,c-diamide synthase | |
| 109 | ORF C109 | 2905378..2905590 [-], 213 | phage-related protein | |
| 110 | PLES_27101 | 2905709..2906455 [-], 747 | hypothetical protein | |
| 111 | PLES_27102 | 2906906..2907406 [+], 501 | hypothetical protein | |
| 112 | PLES_27111 | 2908559..2911444 [+], 2886 | probable sensor/response regulator hybrid | |
| 113 | PLES_27121 | 2911535..2912068 [+], 534 | putative activator of osmoprotectant transporter | |