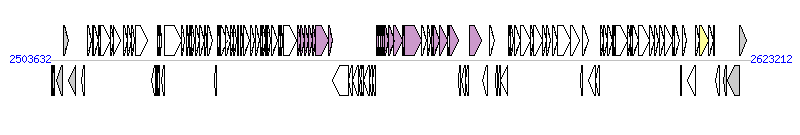The graph information of ICECmeCH34-1 components from CP000352 |
 |
| Complete gene list of ICECmeCH34-1 from CP000352 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | Rmet_6518 | 2503879..2504091 [-], 213 | hypothetical protein | |
| 2 | Rmet_2282 | 2504104..2504430 [-], 327 | conserved hypothetical protein (secreted) | |
| 3 | Rmet_2283 | 2504698..2505786 [-], 1089 | conserved hypothetical protein; putative membrane protein | |
| 4 | yeiE | 2505889..2506806 [+], 918 | putative DNA-binding transcriptional regulator | |
| 5 | Rmet_2285 | 2506827..2508053 [-], 1227 | monovalent cation:proton antiporter (CPA2 family) | |
| 6 | Rmet_2287 | 2509038..2509493 [-], 456 | conserved hypothetical protein; putative membrane protein | |
| 7 | Rmet_2288 | 2509989..2510735 [+], 747 | conserved hypothetical protein | |
| 8 | alpA | 2510854..2511066 [+], 213 | DNA-binding transcriptional activator | |
| 9 | soj | 2511109..2511984 [+], 876 | ATPase involved in plasmid partitioning Soj | |
| 10 | Rmet_2291 | 2511968..2512237 [+], 270 | conserved hypothetical protein | |
| 11 | Rmet_2292 | 2512230..2513909 [+], 1680 | putative regulator | |
| 12 | Rmet_2293 | 2513924..2514484 [+], 561 | putative regulator | |
| 13 | Rmet_2294 | 2514488..2515726 [+], 1239 | conserved hypothetical protein | |
| 14 | Rmet_2295 | 2516156..2516947 [+], 792 | conserved hypothetical protein | |
| 15 | inrR | 2516944..2517471 [+], 528 | Integrase regulator R | |
| 16 | ssb2 | 2517545..2517985 [+], 441 | single-stranded DNA binding protein | |
| 17 | topB2 | 2518269..2520299 [+], 2031 | DNA topoisomerase III | |
| 18 | Rmet_2299 | 2521023..2521658 [-], 636 | Cation efflux system protein | |
| 19 | Rmet_6519 | 2521865..2522035 [-], 171 | hypothetical protein | |
| 20 | Rmet_2300 | 2521983..2522282 [-], 300 | hypothetical protein | |
| 21 | Rmet_6520 | 2522043..2522279 [+], 237 | conserved hypothetical protein | |
| 22 | Rmet_2301 | 2522292..2522666 [+], 375 | hypothetical protein | |
| 23 | pbrR2 | 2522734..2523132 [-], 399 | transcriptional regulator, MerR-family | |
| 24 | cadA | 2523182..2526136 [+], 2955 | lead/cadmium-transporting ATPase (cation efflux ATPase) | |
| 25 | pbrC2 | 2526140..2526640 [+], 501 | prolipoprotein signal peptidase (signal peptidase II) | |
| 26 | Rmet_2305 | 2526977..2527288 [+], 312 | conserved hypothetical protein | |
| 27 | Rmet_2306 | 2527425..2527838 [+], 414 | conserved hypothetical protein | |
| 28 | Rmet_2307 | 2527860..2528252 [+], 393 | conserved hypothetical protein | |
| 29 | Rmet_2308 | 2528435..2529121 [+], 687 | conserved hypothetical protein | |
| 30 | Rmet_2309 | 2529205..2529942 [+], 738 | conserved hypothetical protein | |
| 31 | Rmet_2310 | 2530040..2530318 [+], 279 | conserved hypothetical protein | |
| 32 | Rmet_2311 | 2530607..2531431 [+], 825 | conserved hypothetical protein (protein belongs to CMGI-1) | |
| 33 | merR | 2531693..2532100 [-], 408 | Mercuric resistance operon regulatory protein | |
| 34 | merT | 2532172..2532522 [+], 351 | Mercuric transport protein (Mercury ion transport protein) | |
| 35 | merP | 2532535..2532822 [+], 288 | Mercuric transport protein periplasmic component precursor (Periplasmic mercury ion-binding protein) (Mercury scavenger protein) | |
| 36 | merA' | 2532833..2533951 [+], 1119 | - | |
| 37 | merA'' | 2533662..2534519 [+], 858 | mercuric reductase | |
| 38 | tnpA | 2534538..2535053 [+], 516 | - | |
| 39 | Rmet_2317 | 2534894..2535535 [+], 642 | - | |
| 40 | Rmet_2318 | 2535605..2535865 [+], 261 | conserved hypothetical protein | |
| 41 | Rmet_2319 | 2536173..2536457 [+], 285 | conserved hypothetical protein | |
| 42 | Rmet_2320 | 2536675..2537673 [+], 999 | conserved hypothetical protein | |
| 43 | yubP | 2537819..2538646 [+], 828 | conserved hypothetical protein | |
| 44 | Rmet_2322 | 2538741..2539430 [+], 690 | conserved hypothetical protein | |
| 45 | cggAII | 2539525..2539866 [+], 342 | Conserved plasmid protein | |
| 46 | Rmet_2324 | 2539971..2540378 [+], 408 | conserved hypothetical protein | |
| 47 | Rmet_2325 | 2540395..2540655 [+], 261 | conserved hypothetical protein | |
| 48 | Rmet_2326 | 2540732..2541379 [+], 648 | conserved hypothetical protein | |
| 49 | Rmet_2327 | 2541444..2542553 [+], 1110 | putative adenine-specific DNA methyltransferase | |
| 50 | Rmet_2328 | 2542604..2542924 [+], 321 | conserved hypothetical protein | |
| 51 | Rmet_2329 | 2543014..2543319 [+], 306 | conserved hypothetical protein | |
| 52 | Rmet_2330 | 2543456..2545735 [+], 2280 | Putative plasmid-related DNA/RNA helicase | |
| 53 | pilL | 2545874..2546473 [+], 600 | pilL lipoprotein (type IV pili) | Tfc2, T4SS component |
| 54 | Rmet_2332 | 2546470..2547114 [+], 645 | conserved putative exported protein | Tfc2, T4SS component |
| 55 | Rmet_2333 | 2547127..2547864 [+], 738 | conserved putative exported protein | Tfc3, T4SS component |
| 56 | Rmet_2334 | 2547846..2548436 [+], 591 | putative lytic transglycosylase, catalytic | Tfc4, T4SS component |
| 57 | Rmet_2335 | 2548433..2548981 [+], 549 | putative exported protein | Tfc5, T4SS component |
| 58 | virD4 | 2548986..2551175 [+], 2190 | VirD4, Type IV secretory pathway, VirD4 components | Tfc6, T4SS component |
| 59 | Rmet_2337 | 2551172..2551921 [+], 750 | conserved hypothetical protein; putative membrane protein | Tfc8, T4SS component |
| 60 | Rmet_2338 | 2551962..2554586 [-], 2625 | conserved hypothetical protein | |
| 61 | Rmet_2339 | 2554596..2555321 [-], 726 | conserved hypothetical protein | |
| 62 | Rmet_2340 | 2555360..2556262 [-], 903 | conserved hypothetical protein | |
| 63 | Rmet_2341 | 2556262..2556915 [-], 654 | conserved hypothetical protein | |
| 64 | Rmet_6523 | 2556759..2557229 [-], 471 | conserved hypothetical protein | |
| 65 | Rmet_2342 | 2557226..2558140 [-], 915 | putative AAA ATPase, central region | |
| 66 | Rmet_2343 | 2558158..2558733 [-], 576 | conserved hypothetical protein | |
| 67 | Rmet_2344 | 2558730..2559200 [-], 471 | putative excisionase | |
| 68 | Rmet_2345 | 2559404..2559787 [+], 384 | conserved hypothetical protein | Tfc9, T4SS component |
| 69 | Rmet_2346 | 2559784..2560017 [+], 234 | conserved hypothetical protein | |
| 70 | Rmet_2347 | 2560034..2560393 [+], 360 | conserved hypothetical protein | Tfc10, T4SS component |
| 71 | Rmet_2348 | 2560406..2560804 [+], 399 | conserved hypothetical protein | Tfc11, T4SS component |
| 72 | Rmet_2349 | 2560801..2561493 [+], 693 | conserved hypothetical protein | Tfc12, T4SS component |
| 73 | Rmet_2350 | 2561490..2562401 [+], 912 | conserved hypothetical protein | Tfc13, T4SS component |
| 74 | Rmet_2351 | 2562391..2563809 [+], 1419 | conserved hypothetical protein | Tfc14, T4SS component |
| 75 | Rmet_2352 | 2563790..2564230 [+], 441 | Putative secreted protein | Tfc15, T4SS component |
| 76 | Rmet_2353 | 2564230..2567121 [+], 2892 | Type IV secretory pathway, VirB4 components | Tfc16, T4SS component |
| 77 | Rmet_2354 | 2567146..2567910 [+], 765 | putative exported protein | |
| 78 | radC | 2568086..2568580 [+], 495 | DNA binding protein, possibly associated to lesions | |
| 79 | Rmet_2356 | 2568745..2569191 [+], 447 | conserved hypothetical protein; putative exported protein | Tfc24, T4SS component |
| 80 | Rmet_2357 | 2569188..2570138 [+], 951 | putative exported protein | Tfc23, T4SS component |
| 81 | Rmet_2358 | 2570148..2571542 [+], 1395 | putative membrane protein | Tfc22, T4SS component |
| 82 | Rmet_2359 | 2571539..2571895 [+], 357 | putative membrane protein | |
| 83 | Rmet_2360 | 2571911..2573458 [+], 1548 | conserved hypothetical protein (membrane) | Tfc19, T4SS component |
| 84 | Rmet_2361 | 2573474..2573833 [-], 360 | conserved hypothetical protein | |
| 85 | Rmet_2362 | 2573907..2574539 [-], 633 | conserved hypothetical protein | |
| 86 | Rmet_2363 | 2574552..2575178 [-], 627 | conserved hypothetical protein | |
| 87 | Rmet_2364 | 2575344..2577386 [+], 2043 | relaxase | Relaxase, MOBH Family |
| 88 | Rmet_2365 | 2577434..2578429 [-], 996 | putative transcriptional regulator | |
| 89 | Rmet_2366 | 2578766..2579635 [+], 870 | putative enoyl-CoA hydratase/isomerase | |
| 90 | Rmet_2367 | 2579695..2580120 [-], 426 | conserved hypothetical protein | |
| 91 | Rmet_2368 | 2580177..2580554 [-], 378 | Alkylhydroperoxidase AhpD core | |
| 92 | Rmet_2369 | 2580631..2581857 [-], 1227 | Major facilitator superfamily MFS_1 | |
| 93 | ygaV | 2582007..2582318 [+], 312 | DNA-binding transcriptional regulator | |
| 94 | Rmet_2371 | 2582341..2582868 [+], 528 | Putative detoxifying sulphurtransferase ; inner membrane protein | |
| 95 | Rmet_2372 | 2582975..2583934 [+], 960 | putative hydroxyacylglutathione hydrolase | |
| 96 | Rmet_2373 | 2583983..2585653 [+], 1671 | conserved hypothetical protein | |
| 97 | Rmet_2374 | 2585700..2586179 [+], 480 | hypothetical protein | |
| 98 | ccoN | 2586172..2587728 [+], 1557 | cytochrome-c oxidase | |
| 99 | ccoO | 2587785..2588366 [+], 582 | Cytochrome C oxidase, mono-heme subunit | |
| 100 | Rmet_2377 | 2588507..2589181 [+], 675 | putative cytochrome c4 | |
| 101 | Rmet_2378 | 2589414..2590262 [+], 849 | Putative cytochrome c (fragment) | |
| 102 | ctpA1 | 2590243..2592501 [+], 2259 | metal transporter ATPase | |
| 103 | cutE | 2592526..2594124 [+], 1599 | Apolipoprotein N-acyltransferase | |
| 104 | armS (delta) | 2594278..2594646 [-], 369 | - | |
| 105 | tnpA | 2594664..2595692 [+], 1029 | transposase IS1088 | |
| 106 | armS (delta) | 2595681..2596769 [-], 1089 | - | |
| 107 | armR | 2596766..2597434 [-], 669 | ArmR, DNA-binding response regulator (QseB) | |
| 108 | ccmA | 2597603..2598226 [+], 624 | heme exporter subunit ; ATP-binding component of ABC superfamily | |
| 109 | ccmB | 2598223..2598909 [+], 687 | heme exporter subunit; membrane component of ABC superfamily, cytochrome c maturation protein B | |
| 110 | ccmC | 2598922..2599659 [+], 738 | heme exporter subunit; membrane component of ABC superfamily; protein C | |
| 111 | ccmD | 2599656..2599847 [+], 192 | putative heme exporter protein D,cytochrome c-type biogenesis protein | |
| 112 | ccmE | 2599831..2600277 [+], 447 | periplasmic heme chaperone | |
| 113 | ccmF | 2600281..2602242 [+], 1962 | heme lyase, CcmF subunit | |
| 114 | ccmG | 2602239..2602763 [+], 525 | periplasmic thioredoxin of cytochrome c-type biogenesis | |
| 115 | cycL | 2602760..2603215 [+], 456 | Cytochrome c-type biogenesis protein cycL precursor | |
| 116 | cycH | 2603212..2604093 [+], 882 | putative cytochrome c-type biogenesis protein CycH | |
| 117 | dsbD | 2604191..2606029 [+], 1839 | Thiol:disulfide interchange protein | |
| 118 | dsbE | 2606029..2606865 [+], 837 | putative thiol:disulfide interchange protein | |
| 119 | dsbG | 2606868..2607641 [+], 774 | Putative thiol:disulfide interchange protein | |
| 120 | Rmet_2397 | 2607732..2608538 [+], 807 | putative membrane protein | |
| 121 | Rmet_2398 | 2608581..2609957 [+], 1377 | putative pyridine nucleotide-disulfide oxidoreductase, class I | |
| 122 | Rmet_2399 | 2609989..2610312 [+], 324 | conserved hypothetical protein | |
| 123 | traR2 | 2610350..2611240 [+], 891 | transcriptional regulator | |
| 124 | Rmet_2401 | 2611262..2611555 [-], 294 | Putative HTH-type transcriptional regulator yazB (modular protein) | |
| 125 | traR | 2611675..2612331 [+], 657 | transcriptional regulator | |
| 126 | int | 2612531..2613865 [-], 1335 | - | |
| 127 | tnmB | 2613969..2614538 [+], 570 | protein involved in mobility of Tn6049 | |
| 128 | tnmA | 2614650..2616089 [+], 1440 | transposase Tn6049 | Integrase |
| 129 | gspA | 2616086..2616919 [+], 834 | ATPase (type II general secretion pathway) Tn6049 | |
| 130 | tnmC | 2616973..2617188 [+], 216 | conserved hypothetical protein Tn6049 | |
| 131 | int | 2617367..2617987 [-], 621 | - | |
| 132 | pgsA | 2618628..2619227 [-], 600 | phosphatidylglycerophosphate synthetase | |
| 133 | uvrC | 2619361..2621382 [-], 2022 | excinuclease UvrABC, endonuclease subunit C | |
| 134 | Rmet_2411 | 2621495..2622646 [+], 1152 | conserved hypothetical protein | |