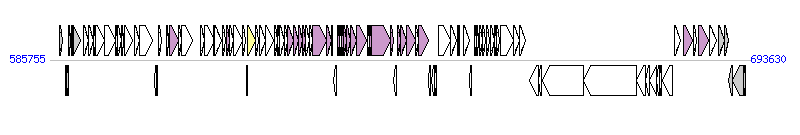The graph information of HAI2 components from BX950851 |
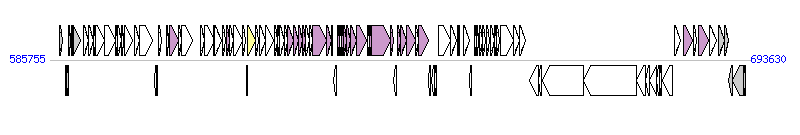 |
| Complete gene list of HAI2 from BX950851 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | ECA0510 | 587151..587651 [+], 501 | putative capsular polysacharide biosynthesis transferase | |
| 2 | ccdA1 | 588090..588308 [-], 219 | putative toxin addiction system protein | |
| 3 | ECA0512 | 588326..588568 [-], 243 | conserved hypothetical protein | |
| 4 | ECA0513 | 588670..588912 [+], 243 | putative phage transcriptional regulator | |
| 5 | ECA0514 | 589060..589218 [+], 159 | putative membrane protein | |
| 6 | ECA0515 | 589365..590507 [+], 1143 | conserved hypothetical protein | |
| 7 | ECA0516 | 590844..591731 [+], 888 | conserved hypothetical protein | |
| 8 | ECA0517 | 591724..592413 [+], 690 | putative membrane protein | |
| 9 | ECA0518 | 592410..592802 [+], 393 | hypothetical protein | |
| 10 | ECA0519 | 592768..594162 [+], 1395 | replicative DNA helicase | |
| 11 | ECA0520 | 594162..595886 [+], 1725 | conserved hypothetical protein | |
| 12 | ECA0521 | 595890..596162 [+], 273 | hypothetical protein | |
| 13 | ECA0522 | 596187..596801 [+], 615 | conserved hypothetical protein | |
| 14 | ECA0522A | 596810..597064 [+], 255 | hypothetical protein | |
| 15 | ECA0523 | 597237..598433 [+], 1197 | conserved hypothetical protein | |
| 16 | ECA0524 | 598803..599600 [+], 798 | conserved hypothetical protein | |
| 17 | topB | 599597..601615 [+], 2019 | putative DNA topoisomerase | |
| 18 | ECA0526 | 601919..602095 [-], 177 | putative phage protein | |
| 19 | ECA0527 | 602138..602293 [-], 156 | hypothetical protein | |
| 20 | ECA0528 | 602464..602955 [+], 492 | conserved hypothetical protein | |
| 21 | ECA0531 | 603654..604091 [+], 438 | putative plasmid-related protein | |
| 22 | pilL | 604237..605553 [+], 1317 | putative Type IV pilus protein | Tfc2, T4SS component |
| 23 | pilM | 605553..605999 [+], 447 | putative Type IV pilus protein | |
| 24 | pilN | 606013..607677 [+], 1665 | putative Type IV pilus protein | |
| 25 | pilP | 608990..609487 [+], 498 | putative Type IV pilus protein | |
| 26 | pilQ | 609487..611046 [+], 1560 | putative Type IV pilus nucleotide-binding protein | |
| 27 | ECA0539 | 611015..611167 [+], 153 | hypothetical protein | |
| 28 | pilR | 611178..612215 [+], 1038 | putative Type IV pilus protein | |
| 29 | pilS | 612271..612876 [+], 606 | putative type IV pilus prepilin | |
| 30 | pilT | 612883..613401 [+], 519 | putative Type IV pilus protein | Orf169_F, T4SS component |
| 31 | pilU | 613398..614057 [+], 660 | putative prepilin peptidase | |
| 32 | pilV | 614071..615465 [+], 1395 | putative Type IV pilus prepilin protein | |
| 33 | ECA0545 | 615469..615948 [+], 480 | alternative C-terminus for the PilV protein (fragment) | |
| 34 | ECA0545A | 615967..616182 [-], 216 | alternative C-terminus for the PilV protein (fragment) | |
| 35 | rci | 616235..617362 [+], 1128 | shufflon-specific DNA recombinase | Integrase |
| 36 | ECA0547 | 617411..617902 [+], 492 | conserved hypothetical protein | |
| 37 | traE | 618090..618890 [+], 801 | putative plasmid transfer protein | |
| 38 | traF | 619034..620236 [+], 1203 | putative plasmid transfer protein | |
| 39 | ECA0550 | 620303..620752 [+], 450 | putative plasmid protein protein | |
| 40 | ECA0551 | 620819..621118 [+], 300 | putative membrane protein | |
| 41 | ECA0552 | 621156..621848 [+], 693 | conserved hypothetical protein | |
| 42 | ECA0553 | 621946..622320 [+], 375 | putative exported protein | |
| 43 | ECA0554 | 622423..623298 [+], 876 | putative membrane protein | Tfc2, T4SS component |
| 44 | ECA0555 | 623308..624021 [+], 714 | putative exported protein | Tfc3, T4SS component |
| 45 | ECA0556 | 624021..624608 [+], 588 | putative exported protein | Tfc4, T4SS component |
| 46 | ECA0557 | 624609..625130 [+], 522 | putative exported protein | Tfc5, T4SS component |
| 47 | ECA0558 | 625127..625744 [+], 618 | putative membrane protein | |
| 48 | ECA0559 | 625722..626204 [+], 483 | putative membrane protein | |
| 49 | ECA0560 | 626208..628304 [+], 2097 | putative plasmid transfer protein | Tfc6, T4SS component |
| 50 | ECA0561 | 628306..629061 [+], 756 | putative membrane protein | Tfc8, T4SS component |
| 51 | ECA0562 | 629084..629278 [+], 195 | hypothetical protein | |
| 52 | ECA0563 | 629420..629959 [-], 540 | putative membrane protein | |
| 53 | ECA0564 | 630094..630414 [+], 321 | putative exported protein | Tfc9, T4SS component |
| 54 | ECA0565 | 630416..630655 [+], 240 | putative membrane protein | |
| 55 | ECA0566 | 630682..631023 [+], 342 | putative membrane protein | Tfc10, T4SS component |
| 56 | ECA0567 | 631033..631395 [+], 363 | putative membrane protein | Tfc11, T4SS component |
| 57 | ECA0568 | 631392..632048 [+], 657 | putative membrane protein | Tfc12, T4SS component |
| 58 | ECA0569 | 632045..632971 [+], 927 | putative exported protein | Tfc13, T4SS component |
| 59 | ECA0570 | 632961..634493 [+], 1533 | putative exported protein | Tfc14, T4SS component |
| 60 | ECA0571 | 634611..634946 [+], 336 | hypothetical protein | |
| 61 | ECA0572 | 634939..635349 [+], 411 | putative lipoprotein | Tfc15, T4SS component |
| 62 | ECA0573 | 635349..638210 [+], 2862 | putative plasmid-related protein | Tfc16, T4SS component |
| 63 | ECA0574 | 638207..638653 [+], 447 | conserved hypothetical protein | Tfc17, T4SS component |
| 64 | ECA0575 | 638755..639189 [-], 435 | putative membrane protein | |
| 65 | ECA0576 | 639329..639733 [+], 405 | putative exported protein | Tfc24, T4SS component |
| 66 | ECA0577 | 639726..640700 [+], 975 | putative exported protein | Tfc23, T4SS component |
| 67 | ECA0578 | 640710..642134 [+], 1425 | putative exported protein | Tfc22, T4SS component |
| 68 | ECA0579 | 642137..642508 [+], 372 | putative exported protein | |
| 69 | ECA0580 | 642505..644037 [+], 1533 | putative membrane protein | Tfc19, T4SS component |
| 70 | ECA0581 | 644070..644459 [-], 390 | putative membrane protein | |
| 71 | ECA0582 | 644602..644961 [-], 360 | putative plasmid-related protein | |
| 72 | ECA0583 | 644958..645260 [-], 303 | putative plasmid-related protein | |
| 73 | ECA0584 | 645558..647429 [+], 1872 | restriction enzyme alpha subunit | |
| 74 | ECA0585 | 647419..648405 [+], 987 | restriction enzyme beta subunit | |
| 75 | ECA0586 | 648538..648678 [+], 141 | hypothetical protein | |
| 76 | ECA0586A | 648731..648898 [+], 168 | hypothetical protein | |
| 77 | ECA0587 | 649514..650404 [+], 891 | conserved hypothetical protein | |
| 78 | ECA0588 | 650412..650702 [-], 291 | putative plasmid-related protein | |
| 79 | ECA0589 | 651097..651408 [+], 312 | hypothetical protein | |
| 80 | ECA0590 | 651458..651814 [+], 357 | hypothetical protein | |
| 81 | ECA0591 | 651896..652369 [+], 474 | hypothetical protein | |
| 82 | ECA0592 | 652458..652862 [+], 405 | hypothetical protein | |
| 83 | ECA0593 | 653073..653486 [+], 414 | putative DNA repair protein | |
| 84 | ECA0594 | 653663..654259 [+], 597 | conserved hypothetical protein | |
| 85 | ECA0595 | 654256..654528 [+], 273 | conserved hypothetical protein | |
| 86 | ECA0596 | 654599..655084 [+], 486 | conserved hypothetical protein | |
| 87 | ECA0597 | 655150..657111 [+], 1962 | conserved hypothetical protein | |
| 88 | ECA0598 | 657186..658079 [+], 894 | conserved hypothetical protein | |
| 89 | ECA0599 | 658154..659074 [+], 921 | conserved hypothetical protein | |
| 90 | cfa8B | 659713..661026 [-], 1314 | putative oxidoreductase | |
| 91 | cfa8A | 661110..661541 [-], 432 | putative oxidoreductase | |
| 92 | cfa7 | 661605..667991 [-], 6387 | type I polyketide synthase | |
| 93 | cfa6 | 667988..676129 [-], 8142 | type I polyketide synthase | |
| 94 | cfa5 | 676143..677582 [-], 1440 | coronafacic acid synthetase, ligase component | |
| 95 | cfa4 | 677579..678112 [-], 534 | coronafacic acid synthetase component | |
| 96 | cfa3 | 678109..679254 [-], 1146 | Cfa-beta-ketoacylsynthase | |
| 97 | cfa2 | 679248..679745 [-], 498 | coronafacic acid dehydratase | |
| 98 | cfa1 | 679742..680017 [-], 276 | Cfa-acyl carrier protein | |
| 99 | cfl | 680119..681681 [-], 1563 | coronafacate ligase | |
| 100 | ECA0610 | 681941..682903 [+], 963 | LysR-family transcriptional regulator | |
| 101 | ECA0611 | 683332..684792 [+], 1461 | conserved hypothetical protein | TraI_F, T4SS component |
| 102 | ECA0612 | 684912..685403 [+], 492 | conserved hypothetical protein | |
| 103 | ECA0613 | 685696..687294 [+], 1599 | conserved hypothetical protein | Relaxase, MOBH Family |
| 104 | ECA0614 | 687380..688402 [+], 1023 | putative phage integrase | |
| 105 | ECA0615 | 688832..689677 [+], 846 | AraC-family transcriptional regulator | |
| 106 | ECA0616 | 689713..690303 [+], 591 | putative membrane protein | |
| 107 | ECA0617 | 690300..690875 [-], 576 | conserved hypothetical protein | |
| 108 | dsbD | 690918..692651 [-], 1734 | thiol:disulfide interchange protein | |
| 109 | cutA | 692627..692962 [-], 336 | periplasmic divalent cation tolerance protein | |