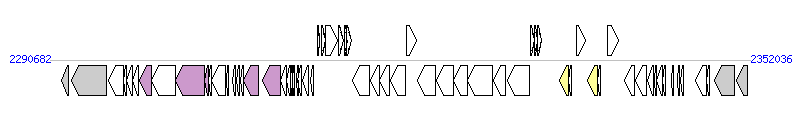The graph information of Tn6098 components from CP001834 |
 |
| Complete gene list of Tn6098 from CP001834 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | yvcA | 2291721..2292314 [-], 594 | Hypothetical protein | |
| 2 | snf | 2292530..2295634 [-], 3105 | DNA/RNA helicase, SWF/SNF family | |
| 3 | LLKF_2229 | 2295829..2297100 [-], 1272 | Integrase, conjugative transposon Tn5276 | |
| 4 | LLKF_2230 | 2297161..2297370 [-], 210 | Excisionase, conjugative transposon Tn5276 | |
| 5 | ardA | 2297429..2297920 [-], 492 | Conjugative transposon antirestriction protein | |
| 6 | LLKF_2232 | 2297944..2298438 [-], 495 | Hypothetical protein | |
| 7 | LLKF_2233 | 2298533..2299597 [-], 1065 | N-acetylmuramoyl-L-alanine amidase, CHAP domain family | PrgK, T4SS component |
| 8 | LLKF_2234 | 2299613..2301658 [-], 2046 | Hypothetical protein | |
| 9 | LLKF_2235 | 2301659..2304226 [-], 2568 | Conjugal transfer protein | YddE, T4SS component |
| 10 | LLKF_2236 | 2304226..2304591 [-], 366 | Hypothetical protein | YddD, T4SS component |
| 11 | LLKF_2237 | 2304600..2304833 [-], 234 | Hypothetical protein | |
| 12 | LLKF_2238 | 2304839..2306107 [-], 1269 | Hypothetical protein | |
| 13 | LLKF_2239 | 2306111..2306311 [-], 201 | Hypothetical protein | |
| 14 | LLKF_2240 | 2306681..2306941 [-], 261 | Hypothetical protein | |
| 15 | LLKF_2241 | 2307055..2307315 [-], 261 | Hypothetical protein | |
| 16 | LLKF_2242 | 2307348..2307614 [-], 267 | Hypothetical protein | |
| 17 | LLKF_2243 | 2307639..2308916 [-], 1278 | Replication initiation factor | |
| 18 | LLKF_2244 | 2309297..2310916 [-], 1620 | DNA segregation ATPase, FtsK/SpoIIIE family | YdcQ, T4SS component |
| 19 | LLKF_2245 | 2310926..2311432 [-], 507 | Hypothetical protein | |
| 20 | LLKF_2246 | 2311435..2311770 [-], 336 | Hypothetical protein | |
| 21 | LLKF_2247 | 2311774..2311896 [-], 123 | Hypothetical protein | |
| 22 | LLKF_2248 | 2311955..2312146 [-], 192 | Hypothetical protein | |
| 23 | LLKF_2249 | 2312285..2312419 [-], 135 | Hypothetical protein | |
| 24 | LLKF_2250 | 2312416..2312682 [-], 267 | Hypothetical protein | |
| 25 | LLKF_2251 | 2312819..2313370 [-], 552 | Hypothetical protein | |
| 26 | LLKF_2252 | 2313480..2313800 [-], 321 | Transcriptional regulator | |
| 27 | LLKF_2253 | 2314169..2314327 [+], 159 | Hypothetical protein | |
| 28 | LLKF_2254 | 2314496..2314840 [+], 345 | Transcriptional regulator | |
| 29 | LLKF_2255 | 2314818..2315900 [+], 1083 | Hypothetical protein | |
| 30 | LLKF_2256 | 2315964..2316416 [+], 453 | IS3/IS911transposase, N-terminal fragment | |
| 31 | LLKF_2257 | 2316470..2316754 [+], 285 | - | |
| 32 | LLKF_2258 | 2316702..2317118 [+], 417 | - | |
| 33 | sucP | 2317224..2318669 [-], 1446 | Sucrose phosphorylase | |
| 34 | LLKF_2260 | 2318756..2319583 [-], 828 | Sugar ABC transporter, permease protein | |
| 35 | LLKF_2261 | 2319596..2320465 [-], 870 | Sugar ABC transporter, permease protein | |
| 36 | LLKF_2262 | 2320566..2321810 [-], 1245 | Sugar ABC transporter, sugar-binding protein | |
| 37 | LLKF_2263 | 2321960..2322829 [+], 870 | Transcriptional regulator, AraC family | |
| 38 | purH | 2322901..2324445 [-], 1545 | phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | |
| 39 | galT | 2324570..2326054 [-], 1485 | Galactose 1-phosphate uridylyltransferase | |
| 40 | galK | 2326051..2327241 [-], 1191 | Galactokinase | |
| 41 | aga | 2327238..2329454 [-], 2217 | Alpha-galactosidase | |
| 42 | galR | 2329585..2330622 [-], 1038 | Transcriptional regulator, LacI family | |
| 43 | fbp | 2330747..2332669 [-], 1923 | Fructose 1,6-bisphosphatase | |
| 44 | LLKF_2270 | 2332809..2333120 [+], 312 | Hypothetical protein, N-terminal fragment | |
| 45 | tnp | 2333148..2333432 [+], 285 | - | |
| 46 | LLKF_2272 | 2333380..2333796 [+], 417 | - | |
| 47 | LLKF_2273 | 2335306..2336160 [-], 855 | IS981 transposase B | Integrase |
| 48 | LLKF_2274 | 2336157..2336417 [-], 261 | IS981 transposase A | |
| 49 | LLKF_2275 | 2336808..2337641 [+], 834 | Hypothetical protein | |
| 50 | LLKF_2276 | 2337802..2338656 [-], 855 | IS981 transposase B | Integrase |
| 51 | LLKF_2277 | 2338653..2338913 [-], 261 | IS981 transposase A | |
| 52 | LLKF_2278 | 2339521..2340483 [+], 963 | D-Isomer specific 2-hydroxyacid dehydrogenase, NAD-dependent | |
| 53 | ceo | 2341013..2341936 [-], 924 | N5-(Carboxyethyl)ornithine synthase | |
| 54 | corA | 2341975..2342925 [-], 951 | Mg2+ and Co2+ transporter, CorA family | |
| 55 | ymgG | 2343050..2343601 [-], 552 | General stress protein, Gls24 family | |
| 56 | ymgH | 2343622..2343810 [-], 189 | Hypothetical protein | |
| 57 | ymgI | 2343820..2344380 [-], 561 | Hypothetical protein | |
| 58 | ymgJ | 2344410..2344649 [-], 240 | Hypothetical protein | |
| 59 | cspB | 2345164..2345364 [-], 201 | cold-shock protein | |
| 60 | cspC | 2345642..2345842 [-], 201 | cold-shock protein | |
| 61 | cspA | 2345973..2346173 [-], 201 | cold-shock protein | |
| 62 | yvcC | 2347228..2348238 [-], 1011 | - | |
| 63 | yvcC | 2348252..2348527 [-], 276 | - | |
| 64 | poxL | 2348944..2350647 [-], 1704 | Pyruvate oxidase | |
| 65 | yvdA | 2350844..2351830 [-], 987 | Beta-lactamase family protein | |