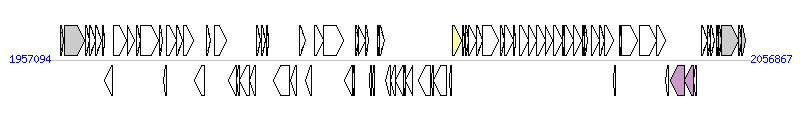The graph information of ICEBvu8482-2 components from CP000139 |
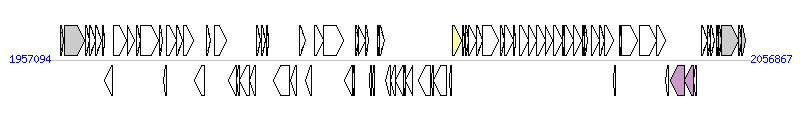 |
| Complete gene list of ICEBvu8482-2 from CP000139 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | BVU_1477 | 1958531..1959019 [+], 489 | hypothetical protein | |
| 2 | BVU_1478 | 1959097..1961955 [+], 2859 | conserved hypothetical protein | |
| 3 | BVU_1479 | 1962094..1962750 [+], 657 | hypothetical protein | |
| 4 | BVU_1480 | 1962695..1963591 [+], 897 | hypothetical protein | |
| 5 | BVU_1481 | 1963599..1964441 [+], 843 | capsular polysaccharide biosynthesis protein D | |
| 6 | BVU_1482 | 1964548..1964925 [+], 378 | hypothetical protein | |
| 7 | BVU_1483 | 1964931..1965995 [-], 1065 | putative peptidase/deacetylase | |
| 8 | BVU_1484 | 1966209..1968014 [+], 1806 | putative long-chain-fatty-acid--CoA ligase | |
| 9 | BVU_1485 | 1968072..1969187 [+], 1116 | peptide chain release factor RF-2 | |
| 10 | BVU_1486 | 1969482..1969952 [+], 471 | conserved hypothetical protein | |
| 11 | BVU_1487 | 1970042..1972516 [+], 2475 | putative TonB-dependent receptor | |
| 12 | BVU_1488 | 1972685..1973188 [+], 504 | RNA polymerase ECF-type sigma factor | |
| 13 | BVU_1489 | 1973269..1973655 [-], 387 | conserved hypothetical protein | |
| 14 | BVU_1490 | 1973751..1975115 [+], 1365 | Na+-dependent transporter | |
| 15 | BVU_1491 | 1975123..1976025 [+], 903 | conserved hypothetical protein | |
| 16 | BVU_1492 | 1976170..1977528 [+], 1359 | putative transposase | |
| 17 | BVU_1493 | 1977712..1979130 [-], 1419 | alkaline phosphatase III precursor | |
| 18 | BVU_1494 | 1979434..1979934 [+], 501 | conserved hypothetical protein | |
| 19 | BVU_1495 | 1980542..1982197 [+], 1656 | transposase | |
| 20 | BVU_1496 | 1982491..1983687 [-], 1197 | conserved hypothetical protein | |
| 21 | BVU_1497 | 1983700..1984047 [-], 348 | excisionase | |
| 22 | BVU_1498 | 1984092..1985477 [-], 1386 | conserved hypothetical protein | |
| 23 | BVU_1499 | 1985555..1986394 [-], 840 | putative transcriptional regulator | |
| 24 | BVU_1500 | 1986505..1987101 [+], 597 | galactoside O-acetyltransferase | |
| 25 | BVU_1501 | 1987262..1987744 [+], 483 | phosphoglycolate phosphatase | |
| 26 | BVU_1502 | 1988016..1988267 [+], 252 | conserved hypothetical protein | |
| 27 | BVU_1503 | 1988890..1991292 [-], 2403 | conserved hypothetical protein | |
| 28 | BVU_1504 | 1991306..1992265 [-], 960 | putative ATPase | |
| 29 | BVU_1505 | 1992589..1993452 [+], 864 | DNA primase | |
| 30 | BVU_1506 | 1993549..1994397 [-], 849 | conserved hypothetical protein | |
| 31 | BVU_1507 | 1994745..1996043 [+], 1299 | predicted ATPase | |
| 32 | BVU_1508 | 1996040..1998910 [+], 2871 | probable helicase | |
| 33 | BVU_1509 | 1999130..2000209 [-], 1080 | hypothetical protein | |
| 34 | BVU_1510 | 2000226..2000552 [-], 327 | hypothetical protein | |
| 35 | BVU_1511 | 2000651..2001001 [+], 351 | hypothetical protein | |
| 36 | BVU_1512 | 2001012..2002016 [+], 1005 | transposase | |
| 37 | BVU_1513 | 2002132..2002506 [+], 375 | hypothetical protein | |
| 38 | BVU_1514 | 2002684..2002983 [-], 300 | conserved hypothetical protein | |
| 39 | BVU_1515 | 2002991..2003284 [-], 294 | conserved hypothetical protein | |
| 40 | BVU_1516 | 2003778..2004038 [+], 261 | conserved hypothetical protein | |
| 41 | BVU_1517 | 2004095..2004799 [+], 705 | conserved hypothetical protein | |
| 42 | BVU_1518 | 2004883..2005674 [-], 792 | ThiF family protein, ubiquitin-activating enzyme | |
| 43 | BVU_1519 | 2005671..2006372 [-], 702 | conserved hypothetical protein | |
| 44 | BVU_1520 | 2006383..2007492 [-], 1110 | conserved hypothetical protein | |
| 45 | BVU_1521 | 2007529..2007750 [-], 222 | conserved hypothetical protein | |
| 46 | BVU_1522 | 2007773..2008828 [-], 1056 | conserved hypothetical protein | |
| 47 | BVU_1523 | 2009590..2011362 [-], 1773 | DNA topoisomerase I | |
| 48 | BVU_1524 | 2011435..2011710 [-], 276 | conserved hypothetical protein | |
| 49 | BVU_1525 | 2011765..2013603 [-], 1839 | conserved hypothetical protein | |
| 50 | BVU_1526 | 2013972..2014262 [-], 291 | hypothetical protein | |
| 51 | BVU_1527 | 2014461..2015684 [+], 1224 | integrase | Integrase |
| 52 | BVU_1528 | 2015942..2016166 [+], 225 | hypothetical protein | |
| 53 | BVU_1529 | 2016260..2016727 [+], 468 | putative transcriptional regulator | |
| 54 | BVU_1530 | 2016752..2017858 [+], 1107 | undecaprenyl-phosphate alpha-N-acetylglucosaminyltransferase | |
| 55 | BVU_1531 | 2017885..2018679 [+], 795 | polysaccharide export protein, BexD/CtrA/VexA family | |
| 56 | BVU_1532 | 2018721..2021087 [+], 2367 | tyrosine-protein kinase ptk | |
| 57 | BVU_1533 | 2021285..2021851 [+], 567 | capsular polysaccharide biosythesis protein, putative | |
| 58 | BVU_1534 | 2021933..2023138 [+], 1206 | putative UDP-ManNAc dehydrogenase | |
| 59 | BVU_1535 | 2023183..2023434 [+], 252 | hypothetical protein | |
| 60 | BVU_1536 | 2023957..2025300 [+], 1344 | putative LPS biosynthesis related polysaccharide transporter/flippase | |
| 61 | BVU_1537 | 2025288..2026490 [+], 1203 | hypothetical protein | |
| 62 | BVU_1538 | 2026497..2027666 [+], 1170 | putative glycosyltransferase | |
| 63 | BVU_1539 | 2027672..2028745 [+], 1074 | putative dehydratase | |
| 64 | BVU_1540 | 2028920..2030074 [+], 1155 | putative epimerase/dehydratase | |
| 65 | BVU_1541 | 2030081..2030479 [+], 399 | conserved hypothetical protein | |
| 66 | BVU_1542 | 2030486..2031667 [+], 1182 | putative UDP-N-acetylglucosamine 2-epimerase | |
| 67 | BVU_1543 | 2031699..2032880 [+], 1182 | hypothetical protein | |
| 68 | BVU_1544 | 2032950..2033288 [+], 339 | hypothetical protein | |
| 69 | BVU_1545 | 2033293..2033673 [+], 381 | putative serine transferase family protein | |
| 70 | BVU_1546 | 2034265..2035458 [+], 1194 | conserved hypothetical protein | |
| 71 | BVU_1547 | 2035471..2036169 [+], 699 | glycosyltransferase family 26 | |
| 72 | BVU_1548 | 2036181..2037341 [+], 1161 | putative UDP-GlcNAc 2-epimerase | |
| 73 | BVU_1549 | 2037432..2037698 [-], 267 | hypothetical protein | |
| 74 | BVU_1550 | 2038237..2038545 [+], 309 | hypothetical protein | |
| 75 | BVU_1551 | 2038570..2040831 [+], 2262 | helicase, putative | |
| 76 | BVU_1552 | 2041149..2043536 [+], 2388 | two-component system sensor histidine kinase | |
| 77 | BVU_1553 | 2043472..2044785 [+], 1314 | putative two-component system response regulator | |
| 78 | BVU_1554 | 2044791..2045207 [-], 417 | conserved hypothetical protein | |
| 79 | BVU_1555 | 2045507..2047543 [-], 2037 | putative mobilization protein | T4CP |
| 80 | BVU_1556 | 2047580..2048827 [-], 1248 | putative mobilization protein | Relaxase, MOBP Family |
| 81 | BVU_1557 | 2048812..2049234 [-], 423 | conserved hypothetical protein | |
| 82 | BVU_1558 | 2049959..2050753 [+], 795 | conserved protein found in conjugate transposon | |
| 83 | BVU_1559 | 2050762..2051211 [+], 450 | conserved hypothetical protein | |
| 84 | BVU_1560 | 2051199..2051867 [+], 669 | conserved hypothetical protein | |
| 85 | BVU_1561 | 2052129..2052431 [+], 303 | conserved protein found in conjugate transposon | |
| 86 | BVU_1562 | 2052437..2052763 [+], 327 | conserved protein found in conjugate transposon | |
| 87 | BVU_1563 | 2052760..2055294 [+], 2535 | conserved protein found in conjugate transposon | |
| 88 | BVU_1564 | 2055291..2055647 [+], 357 | conserved protein found in conjugate transposon | |
| 89 | BVU_1565 | 2055644..2056273 [+], 630 | conserved protein found in conjugate transposon | |