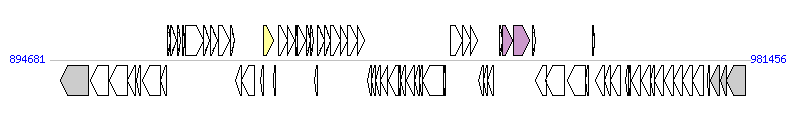The graph information of ICEBvu8482-1 components from CP000139 |
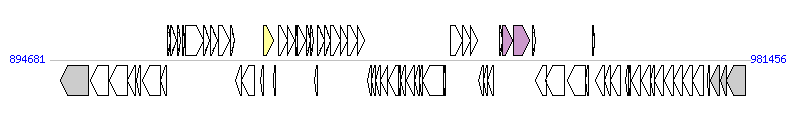 |
| Complete gene list of ICEBvu8482-1 from CP000139 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | BVU_0623 | 895991..899392 [-], 3402 | putative outer membrane protein, probably involved in nutrient binding | |
| 2 | BVU_0624 | 899681..901993 [-], 2313 | glycoside hydrolase family 92 | |
| 3 | BVU_0625 | 902023..904305 [-], 2283 | glycoside hydrolase family 92 | |
| 4 | BVU_0626 | 904323..905315 [-], 993 | putative anti-sigma factor | |
| 5 | BVU_0627 | 905344..905919 [-], 576 | RNA polymerase ECF-type sigma factor | |
| 6 | BVU_0628 | 906124..908346 [-], 2223 | glycoside hydrolase family 92 | |
| 7 | BVU_0629 | 908440..909135 [-], 696 | hypothetical protein | |
| 8 | BVU_0630 | 909295..909573 [+], 279 | conserved hypothetical protein | |
| 9 | BVU_0631 | 909582..910397 [+], 816 | transcriptional regulator | |
| 10 | BVU_0632 | 910471..910878 [+], 408 | putative general stress protein | |
| 11 | BVU_0633 | 911107..911415 [+], 309 | conserved hypothetical protein | |
| 12 | BVU_0634 | 911433..913661 [+], 2229 | ATP-dependent Clp protease, ATP-binding subunit ClpA | |
| 13 | BVU_0635 | 913757..914410 [+], 654 | Leu/Phe-tRNA-protein transferase | |
| 14 | BVU_0636 | 914551..915390 [+], 840 | hypothetical protein | |
| 15 | BVU_0637 | 915605..916897 [+], 1293 | BexA, putative cation effux pump | |
| 16 | BVU_0638 | 917012..917596 [+], 585 | conserved hypothetical protein | |
| 17 | BVU_0639 | 917614..918384 [-], 771 | transcriptional regulator | |
| 18 | BVU_0640 | 918435..920042 [-], 1608 | glycoside hydrolase family 27 | |
| 19 | BVU_0643 | 920743..921141 [-], 399 | hypothetical protein | |
| 20 | BVU_0644 | 921126..922361 [+], 1236 | transposase | Integrase |
| 21 | BVU_0645 | 922442..922636 [-], 195 | hypothetical protein | |
| 22 | BVU_0646 | 922956..924083 [+], 1128 | conserved hypothetical protein | |
| 23 | BVU_0647 | 924087..924980 [+], 894 | conserved hypothetical protein | |
| 24 | BVU_0648 | 925056..925421 [+], 366 | hypothetical protein | |
| 25 | BVU_0649 | 925418..926398 [+], 981 | conserved hypothetical protein | |
| 26 | BVU_0650 | 926441..926920 [+], 480 | conserved hypothetical protein | |
| 27 | BVU_0651 | 927015..927305 [+], 291 | conserved hypothetical protein | |
| 28 | BVU_0652 | 927455..927850 [-], 396 | hypothetical protein | |
| 29 | BVU_0653 | 927852..928658 [+], 807 | conserved hypothetical protein | |
| 30 | BVU_0654 | 928660..929418 [+], 759 | type I restriction enzyme, M subunit | |
| 31 | BVU_0655 | 929437..930666 [+], 1230 | hypothetical protein | |
| 32 | BVU_0656 | 930679..931560 [+], 882 | conserved hypothetical protein | |
| 33 | BVU_0657 | 931587..932741 [+], 1155 | conserved hypothetical protein | |
| 34 | BVU_0658 | 932804..933715 [+], 912 | hypothetical protein | |
| 35 | BVU_0659 | 934077..934556 [-], 480 | glycoside hydrolase family 24 | |
| 36 | BVU_0660 | 934553..935011 [-], 459 | conserved protein found in conjugate transposon | |
| 37 | BVU_0661 | 935032..935619 [-], 588 | conserved protein found in conjugate transposon | |
| 38 | BVU_0662 | 935616..936530 [-], 915 | conserved protein found in conjugate transposon | |
| 39 | BVU_0663 | 936581..937888 [-], 1308 | conserved protein found in conjugate transposon | |
| 40 | BVU_0664 | 937839..938141 [-], 303 | conserved hypothetical protein | |
| 41 | BVU_0665 | 938138..938761 [-], 624 | conserved protein found in conjugate transposon | |
| 42 | BVU_0666 | 938884..939897 [-], 1014 | conserved protein found in conjugate transposon | |
| 43 | BVU_0667 | 939900..940529 [-], 630 | conserved protein found in conjugate transposon | |
| 44 | BVU_0668 | 940526..940894 [-], 369 | conserved protein found in conjugate transposon | |
| 45 | BVU_0669 | 940891..943416 [-], 2526 | conserved protein found in conjugate transposon | |
| 46 | BVU_0670 | 943413..943742 [-], 330 | conserved protein found in conjugate transposon | |
| 47 | BVU_0671 | 944349..945782 [+], 1434 | conserved hypothetical protein | |
| 48 | BVU_0672 | 945769..946758 [+], 990 | conserved hypothetical protein | |
| 49 | BVU_0673 | 946755..947660 [+], 906 | hypothetical protein | |
| 50 | BVU_0674 | 947775..948440 [-], 666 | conserved hypothetical protein | |
| 51 | BVU_0675 | 948428..948877 [-], 450 | conserved hypothetical protein | |
| 52 | BVU_0676 | 948886..949680 [-], 795 | conserved protein found in conjugate transposon | |
| 53 | BVU_0677 | 950405..950827 [+], 423 | conserved hypothetical protein | |
| 54 | BVU_0678 | 950812..952059 [+], 1248 | putative mobilization protein | Relaxase, MOBP Family |
| 55 | BVU_0679 | 952096..954132 [+], 2037 | putative mobilization protein | T4CP |
| 56 | BVU_0680 | 954432..954848 [+], 417 | conserved hypothetical protein | |
| 57 | BVU_0681 | 954854..956167 [-], 1314 | putative two-component system response regulator | |
| 58 | BVU_0682 | 956103..958490 [-], 2388 | two-component system sensor histidine kinase | |
| 59 | BVU_0683 | 958808..961069 [-], 2262 | helicase, putative | |
| 60 | BVU_0684 | 961094..961402 [-], 309 | hypothetical protein | |
| 61 | BVU_0685 | 961941..962207 [+], 267 | hypothetical protein | |
| 62 | BVU_0686 | 962298..963458 [-], 1161 | putative UDP-GlcNAc 2-epimerase | |
| 63 | BVU_0687 | 963470..964168 [-], 699 | glycosyltransferase family 26 | |
| 64 | BVU_0688 | 964181..965374 [-], 1194 | conserved hypothetical protein | |
| 65 | BVU_0689 | 965966..966346 [-], 381 | putative serine acetyltransferase family protein | |
| 66 | BVU_0690 | 966351..966689 [-], 339 | hypothetical protein | |
| 67 | BVU_0691 | 966759..967940 [-], 1182 | hypothetical protein | |
| 68 | BVU_0692 | 967972..969153 [-], 1182 | putative UDP-N-acetylglucosamine 2-epimerase | |
| 69 | BVU_0693 | 969160..969558 [-], 399 | conserved hypothetical protein | |
| 70 | BVU_0694 | 969565..970719 [-], 1155 | putative epimerase/dehydratase, involved in capsular polysaccharide biosynthesis | |
| 71 | BVU_0695 | 970894..971967 [-], 1074 | putative dehydratase | |
| 72 | BVU_0696 | 971973..973142 [-], 1170 | putative glycosyltransferase | |
| 73 | BVU_0697 | 973149..974351 [-], 1203 | hypothetical protein | |
| 74 | BVU_0698 | 974339..975682 [-], 1344 | putative LPS biosynthesis related polysaccharide transporter/flippase | |
| 75 | BVU_0699 | 976205..976456 [-], 252 | hypothetical protein | |
| 76 | BVU_0700 | 976501..977706 [-], 1206 | putative UDP-ManNAc dehydrogenase | |
| 77 | BVU_0701 | 977788..978492 [-], 705 | capsular polysaccharide biosythesis protein, putative | |
| 78 | BVU_0702 | 978506..980884 [-], 2379 | tyrosine-protein kinase ptk | |