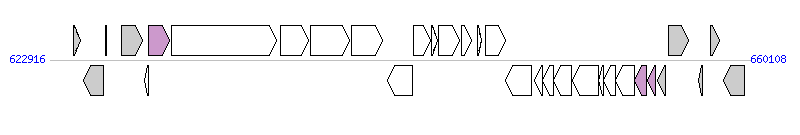The graph information of CTnPg2 components from AP009380 |
 |
| Complete gene list of CTnPg2 from AP009380 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | PGN_0574 | 624175..624516 [+], 342 | conserved hypothetical protein | |
| 2 | PGN_0575 | 624688..625749 [-], 1062 | transposase in ISPg1 | |
| 3 | PGN_0576 | 625842..625943 [+], 102 | hypothetical protein | |
| 4 | PGN_0577 | 626732..627817 [+], 1086 | transposase in ISPg1 | |
| 5 | PGN_0578 | 627916..628137 [-], 222 | conserved hypothetical protein found in conjugate transposon | |
| 6 | PGN_0579 | 628159..629265 [+], 1107 | conserved hypothetical protein | T4CP |
| 7 | PGN_0580 | 629353..634965 [+], 5613 | conserved hypothetical protein | |
| 8 | PGN_0581 | 635151..636635 [+], 1485 | conserved hypothetical protein | |
| 9 | PGN_0582 | 636731..638812 [+], 2082 | DNA topoisomerase I | |
| 10 | PGN_0583 | 638954..640594 [+], 1641 | conserved hypothetical protein | |
| 11 | PGN_0584 | 640869..642197 [-], 1329 | conserved hypothetical protein | |
| 12 | PGN_0585 | 642231..643133 [+], 903 | transposase in ISPg3 | |
| 13 | PGN_0586 | 643172..643486 [+], 315 | conserved hypothetical protein | |
| 14 | PGN_0587 | 643570..644655 [+], 1086 | transposase in ISPg1 | |
| 15 | PGN_0588 | 644756..645304 [+], 549 | conserved hypothetical protein | |
| 16 | PGN_0589 | 645609..645830 [+], 222 | conserved hypothetical protein | |
| 17 | PGN_0590 | 646047..647105 [+], 1059 | putative Fic family protein | |
| 18 | PGN_0591 | 647136..648518 [-], 1383 | conserved hypothetical protein | |
| 19 | traQ | 648638..649075 [-], 438 | putative conserved protein found in conjugate transposon TraQ | |
| 20 | traO | 649095..649679 [-], 585 | putative conserved protein found in conjugate transposon TraO | |
| 21 | traN | 649684..650619 [-], 936 | conserved protein found in conjugate transposon TraN | |
| 22 | traM | 650693..652054 [-], 1362 | putative conserved protein found in conjugate transposon TraM | |
| 23 | PGN_0596 | 652051..652314 [-], 264 | conserved hypothetical protein found in conjugate transposon | |
| 24 | traK | 652318..652941 [-], 624 | putative conserved protein found in conjugate transposon TraK | |
| 25 | traJ | 652959..653945 [-], 987 | conserved transmembrane protein found in conjugate transposon TraJ | |
| 26 | traI | 653966..654589 [-], 624 | putative conserved protein found in conjugate transposon TraI | TrbJ, T4SS component |
| 27 | PGN_0600 | 654641..655108 [-], 468 | conserved hypothetical protein | TraC_F, T4SS component |
| 28 | PGN_0601 | 655186..655605 [-], 420 | conserved hypothetical protein | |
| 29 | PGN_0602 | 655769..656854 [+], 1086 | transposase in ISPg1 | |
| 30 | PGN_0603 | 657387..657560 [-], 174 | hypothetical protein | |
| 31 | PGN_0604 | 657990..658472 [+], 483 | ferritin | |
| 32 | PGN_0605 | 658720..659805 [-], 1086 | transposase in ISPg1 | |