The graph information of CTnPg1-a components from AP009380 |
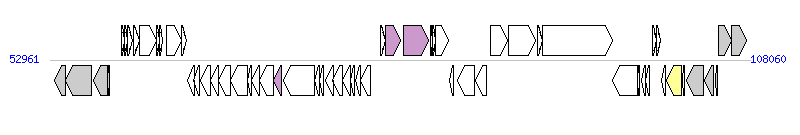 |
| Complete gene list of CTnPg1-a from AP009380 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | PGN_0042 | 53288..54142 [-], 855 | probable phosphatidate cytidylyltransferase | |
| 2 | PGN_0043 | 54171..56192 [-], 2022 | putative transmembrane AAA-metalloprotease FtsH | |
| 3 | PGN_0044 | 56393..57496 [-], 1104 | GTP-binding protein | |
| 4 | PGN_0045 | 57515..57679 [-], 165 | conserved hypothetical protein | |
| 5 | PGN_0046 | 58549..58815 [+], 267 | hypothetical protein | |
| 6 | PGN_0047 | 58828..59046 [+], 219 | conserved hypothetical protein | |
| 7 | PGN_0048 | 59059..59475 [+], 417 | conserved hypothetical protein | |
| 8 | PGN_0049 | 59497..60027 [+], 531 | probable anti-restriction protein | |
| 9 | PGN_0050 | 60044..61318 [+], 1275 | conserved hypothetical protein | |
| 10 | PGN_0051 | 61345..61599 [+], 255 | conserved hypothetical protein | |
| 11 | PGN_0052 | 61604..61834 [+], 231 | hypothetical protein | |
| 12 | PGN_0053 | 62092..63318 [+], 1227 | conserved hypothetical protein | |
| 13 | PGN_0054 | 63320..63724 [+], 405 | hypothetical protein | |
| 14 | PGN_0055 | 63785..64300 [-], 516 | probable lysozyme | |
| 15 | PGN_0056 | 64284..64745 [-], 462 | probable conserved protein found in conjugate transposon | |
| 16 | traP | 64770..65624 [-], 855 | probable conserved protein found in conjugate transposon TraP | |
| 17 | PGN_0058 | 65624..66208 [-], 585 | probable conserved protein found in conjugate transposon | |
| 18 | traN | 66210..67130 [-], 921 | conserved protein found in conjugate transposon TraN | |
| 19 | traM | 67169..68536 [-], 1368 | conserved protein found in conjugate transposon TraM | |
| 20 | PGN_0061 | 68487..68819 [-], 333 | hypothetical protein | |
| 21 | traK | 68816..69439 [-], 624 | putative conserved protein found in conjugate transposon TraK | |
| 22 | traJ | 69459..70535 [-], 1077 | conserved transmembrane protein found in conjugate transposon TraJ | |
| 23 | traI | 70538..71167 [-], 630 | putative conserved protein found in conjugate transposon TraI | TrbJ, T4SS component |
| 24 | traG | 71302..73803 [-], 2502 | conserved protein found in conjugate transposon TraG | |
| 25 | traF | 73800..74177 [-], 378 | probable conserved transmembrane protein found in conjugate transposon TraF | |
| 26 | traF | 74182..74481 [-], 300 | probable conserved transmembrane protein found in conjugate transposon TraE | |
| 27 | PGN_0068 | 74657..75244 [-], 588 | hypothetical protein | |
| 28 | traA | 75232..75972 [-], 741 | probable conserved protein found in conjugate transposon TraA | |
| 29 | PGN_0070 | 76002..76592 [-], 591 | hypothetical protein | |
| 30 | PGN_0071 | 76589..76942 [-], 354 | hypothetical protein | |
| 31 | PGN_0072 | 76968..77399 [-], 432 | hypothetical protein | |
| 32 | traA | 77383..78165 [-], 783 | putative conserved protein found in conjugate transposon TraA | |
| 33 | PGN_0074 | 78954..79367 [+], 414 | conserved hypothetical protein | |
| 34 | PGN_0075 | 79352..80587 [+], 1236 | conserved hypothetical protein | Relaxase, MOBP Family |
| 35 | PGN_0076 | 80770..82779 [+], 2010 | putative mobilization protein TraG family | T4CP |
| 36 | PGN_0077 | 82878..83033 [+], 156 | hypothetical protein | |
| 37 | PGN_0078 | 83075..83287 [+], 213 | hypothetical protein | |
| 38 | PGN_0079 | 83317..84354 [+], 1038 | conserved hypothetical protein with DUF1016 domain | |
| 39 | PGN_0080 | 84397..84723 [-], 327 | probable tetracycline resistance element mobilization regulatory protein RteC | |
| 40 | PGN_0081 | 85010..86368 [-], 1359 | putative Na driven multidrug efflux pump | |
| 41 | PGN_0082 | 86465..87313 [-], 849 | probable transcriptional regulator AraC family | |
| 42 | PGN_0083 | 87598..88992 [+], 1395 | conserved hypothetical protein | |
| 43 | PGN_0084 | 89027..91156 [+], 2130 | DNA topoisomerase I | |
| 44 | PGN_0085 | 91296..91727 [+], 432 | hypothetical protein | |
| 45 | PGN_0086 | 91714..97200 [+], 5487 | putative DNA methylase | |
| 46 | PGN_0087 | 97257..99167 [-], 1911 | conserved hypothetical protein | |
| 47 | PGN_0088 | 99167..99376 [-], 210 | putative transcriptional regulator | |
| 48 | PGN_0089 | 99529..99849 [-], 321 | hypothetical protein | |
| 49 | PGN_0090 | 99833..100186 [-], 354 | hypothetical protein | |
| 50 | PGN_0091 | 100407..100685 [+], 279 | hypothetical protein | |
| 51 | PGN_0092 | 100714..101025 [+], 312 | conserved hypothetical protein | |
| 52 | PGN_0093 | 101075..101440 [-], 366 | conserved hypothetical protein | |
| 53 | PGN_0094 | 101455..102690 [-], 1236 | putative bacteriophage integrase | Integrase |
| 54 | PGN_0095 | 102733..102882 [-], 150 | hypothetical protein | |
| 55 | PGN_0096 | 103084..104424 [-], 1341 | aspartate kinase | |
| 56 | PGN_0097 | 104442..105182 [-], 741 | putative cell-division ATP-binding protein | |
| 57 | PGN_0098 | 105346..105519 [-], 174 | hypothetical protein | |
| 58 | PGN_0099 | 105575..106588 [+], 1014 | probable peptidase | |
| 59 | PGN_0100 | 106598..107746 [+], 1149 | diaminopimelate decarboxylase | |

