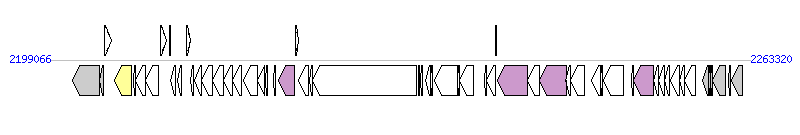The graph information of ICEEfaV583-1 components from AE016830 |
 |
| Complete gene list of ICEEfaV583-1 from AE016830 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | EF_2280 | 2201166..2203613 [-], 2448 | conserved hypothetical protein | |
| 2 | EF_2281 | 2203600..2203989 [-], 390 | conserved hypothetical protein | |
| 3 | EF_2282 | 2204066..2204680 [+], 615 | conserved domain protein | |
| 4 | EF_2283 | 2204960..2206573 [-], 1614 | site-specific recombinase, resolvase family, putative | Integrase |
| 5 | EF_2284 | 2206701..2206877 [-], 177 | hypothetical protein | |
| 6 | EF_2285 | 2206945..2207805 [-], 861 | hypothetical protein | |
| 7 | EF_2286 | 2207802..2209013 [-], 1212 | ParB-like nuclease domain protein | |
| 8 | EF_2287 | 2209172..2209732 [+], 561 | hypothetical protein | |
| 9 | EF_2288 | 2209998..2210117 [+], 120 | hypothetical protein | |
| 10 | EF_2289 | 2210145..2210591 [-], 447 | hypothetical protein | |
| 11 | EF_2290 | 2210747..2211127 [-], 381 | RNA polymerase sigma-70 factor, ECF subfamily | |
| 12 | EF_2291 | 2211601..2211966 [+], 366 | transcriptional regulator, Cro/CI family | |
| 13 | EF_2292 | 2211933..2212277 [-], 345 | hypothetical protein | |
| 14 | vanX | 2212353..2212961 [-], 609 | D-alanyl-D-alanine dipeptidase | |
| 15 | vanB | 2212967..2213995 [-], 1029 | D-alanine--D-lactate ligase | |
| 16 | EF_2295 | 2213988..2214959 [-], 972 | D-specific alpha-keto acid dehydrogenase | |
| 17 | vanW | 2214956..2215783 [-], 828 | vancomycin B-type resistance protein VanW | |
| 18 | vanYB | 2215801..2216607 [-], 807 | D-alanyl-D-alanine carboxypeptidase | |
| 19 | vanSB | 2216783..2218126 [-], 1344 | sensor histidine kinase VanSB | |
| 20 | vanRB | 2218126..2218788 [-], 663 | DNA-binding response regulator VanRB | |
| 21 | EF_2300 | 2218843..2219031 [-], 189 | streptomycin resistance protein, putative | |
| 22 | EF_2302 | 2219565..2219783 [-], 219 | conserved hypothetical protein | |
| 23 | EF_2303 | 2220066..2221478 [-], 1413 | conserved hypothetical protein | Relaxase, MOBP Family |
| 24 | EF_2304 | 2221559..2221834 [+], 276 | transcriptional regulator, Cro/CI family | |
| 25 | EF_2305 | 2221837..2222787 [-], 951 | toprim domain protein | |
| 26 | EF_2306 | 2222854..2223192 [-], 339 | conserved hypothetical protein | |
| 27 | EF_2307 | 2223189..2232710 [-], 9522 | conserved hypothetical protein | |
| 28 | EF_2308 | 2232905..2233006 [-], 102 | hypothetical protein | |
| 29 | EF_2309 | 2233047..2233265 [-], 219 | hypothetical protein | |
| 30 | EF_2310 | 2233538..2233987 [-], 450 | hypothetical protein | |
| 31 | EF_2311 | 2233984..2234145 [-], 162 | hypothetical protein | |
| 32 | topB-2 | 2234375..2236492 [-], 2118 | DNA topoisomerase III | |
| 33 | EF_2313 | 2236443..2236670 [-], 228 | hypothetical protein | |
| 34 | EF_2314 | 2236670..2237956 [-], 1287 | bacteriocin, putative | |
| 35 | EF_2315 | 2238911..2239159 [-], 249 | hypothetical protein | |
| 36 | EF_2316 | 2239156..2240004 [-], 849 | conserved domain protein | |
| 37 | EF_2317 | 2239989..2240090 [+], 102 | hypothetical protein | |
| 38 | EF_2318 | 2240101..2242878 [-], 2778 | peptidase, M23/M37 family | Orf14_Tn, T4SS component |
| 39 | EF_2319 | 2242875..2243990 [-], 1116 | hypothetical protein | |
| 40 | EF_2320 | 2244004..2246481 [-], 2478 | traE protein, putative | PrgJ, T4SS component |
| 41 | EF_2321 | 2246405..2246824 [-], 420 | hypothetical protein | |
| 42 | EF_2322 | 2246825..2248129 [-], 1305 | conserved domain protein | |
| 43 | EF_2324 | 2248728..2249597 [-], 870 | conserved hypothetical protein | |
| 44 | EF_2325 | 2249591..2249782 [-], 192 | hypothetical protein | |
| 45 | EF_2326 | 2249859..2251745 [-], 1887 | group II intron reverse transcriptase maturase | |
| 46 | EF_2327 | 2252465..2252596 [-], 132 | hypothetical protein | |
| 47 | EF_2328 | 2252661..2254436 [-], 1776 | TraG family protein | T4CP |
| 48 | EF_2329 | 2254433..2254912 [-], 480 | hypothetical protein | |
| 49 | EF_2330 | 2254941..2255447 [-], 507 | hypothetical protein | |
| 50 | EF_2331 | 2255485..2255916 [-], 432 | hypothetical protein | |
| 51 | EF_2332 | 2255995..2256852 [-], 858 | conserved domain protein | |
| 52 | EF_2333 | 2256825..2257283 [-], 459 | hypothetical protein | |
| 53 | EF_2334 | 2257334..2258320 [-], 987 | conserved domain protein | |
| 54 | EF_2335 | 2258996..2259496 [-], 501 | conserved hypothetical protein | |
| 55 | EF_2336 | 2259509..2259730 [-], 222 | conserved hypothetical protein | |
| 56 | EF_2337 | 2259735..2259872 [-], 138 | hypothetical protein | |
| 57 | EF_2338 | 2259869..2261053 [-], 1185 | transcriptional regulator, Cro/CI family | |
| 58 | EF_2339 | 2261311..2261565 [-], 255 | hypothetical protein | |
| 59 | EF_2340 | 2261591..2262586 [-], 996 | C-5 cytosine-specific DNA methylase | |