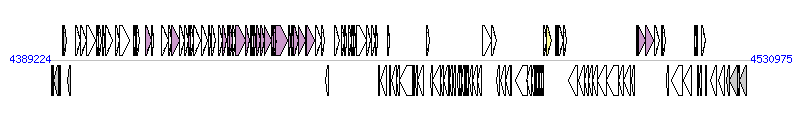The graph information of ICESenTy2-1 components from AE014613 |
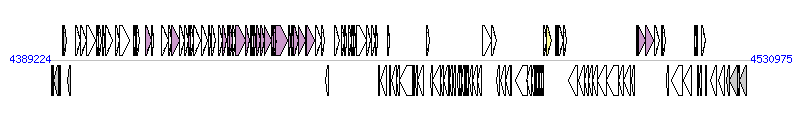 |
| Complete gene list of ICESenTy2-1 from AE014613 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | t4219 | 4389499..4389696 [-], 198 | hypothetical protein | |
| 2 | t4220 | 4389719..4390006 [-], 288 | putative GerE family regulatory protein | |
| 3 | t4221 | 4390003..4390878 [-], 876 | araC family regulatory protein | |
| 4 | t4222 | 4391144..4391362 [-], 219 | hypothetical protein | |
| 5 | t4223 | 4391683..4391976 [+], 294 | conserved hypothetical protein | |
| 6 | t4224 | 4391973..4392464 [+], 492 | putative acetyltransferase | |
| 7 | phoN | 4392705..4393457 [-], 753 | nonspecific acid phosphatase precursor | |
| 8 | t4226 | 4394302..4395336 [+], 1035 | hypothetical protein | |
| 9 | t4227 | 4395333..4396697 [+], 1365 | DNA helicase | |
| 10 | t4228 | 4396690..4398489 [+], 1800 | hypothetical protein | |
| 11 | t4229 | 4398658..4398963 [+], 306 | hypothetical protein | |
| 12 | t4230 | 4399091..4399672 [+], 582 | hypothetical protein | |
| 13 | t4231 | 4399757..4400314 [+], 558 | hypothetical protein | |
| 14 | t4232 | 4400559..4401902 [+], 1344 | hypothetical protein | |
| 15 | t4233 | 4402488..4403267 [+], 780 | hypothetical protein | |
| 16 | topB | 4403378..4405372 [+], 1995 | topoisomerase B | |
| 17 | t4235 | 4406040..4406489 [+], 450 | hypothetical protein | |
| 18 | t4236 | 4406570..4406788 [+], 219 | hypothetical protein | |
| 19 | ssb | 4406801..4407337 [+], 537 | single strand binding protein | |
| 20 | pilL | 4408518..4409762 [+], 1245 | putative exported protein | Tfc2, T4SS component |
| 21 | pilM | 4409765..4410184 [+], 420 | putative exported protein | |
| 22 | pilO | 4411887..4413182 [+], 1296 | putative pilus assembly protein | |
| 23 | pilP | 4413280..4413750 [+], 471 | pilus assembly protein | |
| 24 | pilQ | 4413760..4415475 [+], 1716 | nucleotide-binding protein | TraJ_I, T4SS component |
| 25 | pilR | 4415472..4416557 [+], 1086 | putative membrane protein | |
| 26 | pilS | 4416557..4417192 [+], 636 | prepilin | |
| 27 | pilT | 4417202..4417678 [+], 477 | putative membrane protein | Orf169_F, T4SS component |
| 28 | pilU | 4417675..4418337 [+], 663 | prepilin peptidase | |
| 29 | pilV | 4418334..4419641 [+], 1308 | prepilin | |
| 30 | rci | 4419989..4421140 [+], 1152 | shufflon-specific DNA recombinase | |
| 31 | pilK | 4421237..4421704 [+], 468 | putative exported protein | |
| 32 | t4254 | 4421916..4422791 [+], 876 | hypothetical protein | |
| 33 | t4255 | 4423298..4424212 [+], 915 | putative membrane protein | |
| 34 | t4256 | 4424231..4424971 [+], 741 | putative exported protein | Tfc3, T4SS component |
| 35 | t4257 | 4425085..4425636 [+], 552 | hypothetical protein | Tfc4, T4SS component |
| 36 | t4258 | 4425672..4426172 [+], 501 | hypothetical protein | Tfc5, T4SS component |
| 37 | t4259 | 4426182..4426751 [+], 570 | putative membrane protein | |
| 38 | t4260 | 4426771..4428861 [+], 2091 | hypothetical protein | Tfc6, T4SS component |
| 39 | t4261 | 4428858..4429616 [+], 759 | putative membrane protein | Tfc8, T4SS component |
| 40 | t4262 | 4429835..4430107 [+], 273 | hypothetical protein | Tfc9, T4SS component |
| 41 | t4263 | 4430107..4430346 [+], 240 | putative membrane protein | |
| 42 | t4264 | 4430376..4430738 [+], 363 | putative membrane protein | Tfc10, T4SS component |
| 43 | t4265 | 4431119..4431772 [+], 654 | possible exported protein | Tfc12, T4SS component |
| 44 | t4266 | 4431772..4432671 [+], 900 | hypothetical protein | Tfc13, T4SS component |
| 45 | t4268 | 4432661..4434139 [+], 1479 | putative exported protein | Tfc14, T4SS component |
| 46 | t4267 | 4434132..4434575 [+], 444 | putative lipoprotein | Tfc15, T4SS component |
| 47 | t4269 | 4434572..4434991 [+], 420 | hypothetical protein | Tfc16, T4SS component |
| 48 | t4270 | 4434988..4437411 [+], 2424 | hypothetical protein | Tfc16, T4SS component |
| 49 | t4271 | 4437566..4437952 [+], 387 | hypothetical protein | Tfc17, T4SS component |
| 50 | t4272 | 4438107..4438499 [+], 393 | hypothetical protein | Tfc24, T4SS component |
| 51 | t4273 | 4438496..4439464 [+], 969 | hypothetical protein | Tfc23, T4SS component |
| 52 | t4274 | 4439479..4440909 [+], 1431 | hypothetical protein | Tfc22, T4SS component |
| 53 | t4275 | 4440906..4441238 [+], 333 | putative membrane protein | |
| 54 | t4276 | 4441242..4442747 [+], 1506 | putative membrane protein | Tfc19, T4SS component |
| 55 | t4277 | 4442986..4444233 [+], 1248 | conserved hypothetical protein | |
| 56 | t4278 | 4444245..4444859 [+], 615 | conserved hypothetical protein | |
| 57 | t4279 | 4445070..4445612 [-], 543 | hypothetical protein | |
| 58 | t4280 | 4446878..4447798 [+], 921 | hypothetical protein | |
| 59 | t4281 | 4448315..4448824 [+], 510 | hypothetical protein | |
| 60 | t4282 | 4448920..4449300 [+], 381 | hypothetical protein | |
| 61 | t4283 | 4449766..4450350 [+], 585 | hypothetical protein | |
| 62 | t4284 | 4450347..4450643 [+], 297 | hypothetical protein | |
| 63 | t4285 | 4450721..4451179 [+], 459 | hypothetical protein | |
| 64 | t4286 | 4451268..4453217 [+], 1950 | hypothetical protein | |
| 65 | t4287 | 4453289..4454197 [+], 909 | hypothetical protein | |
| 66 | t4288 | 4454271..4455170 [+], 900 | hypothetical protein | |
| 67 | t4289 | 4455212..4455571 [+], 360 | conserved hypothetical protein | |
| 68 | t4290 | 4455671..4455940 [-], 270 | hypothetical protein | |
| 69 | samB | 4456072..4457346 [-], 1275 | UV protection protein | |
| 70 | t4293 | 4457565..4457942 [+], 378 | hypothetical protein | |
| 71 | t4294 | 4458029..4458247 [-], 219 | putative positive regulator of late gene transcription | |
| 72 | t4295 | 4458315..4459415 [-], 1101 | putative regulator of late gene expression | |
| 73 | t4296 | 4459412..4459897 [-], 486 | putative phage tail protein | |
| 74 | t4297 | 4459897..4462677 [-], 2781 | hypothetical protein | |
| 75 | t4298 | 4462670..4462789 [-], 120 | hypothetical protein | |
| 76 | t4299 | 4462804..4463106 [-], 303 | putative phage tail protein | |
| 77 | t4300 | 4463161..4463676 [-], 516 | probable major tail tube protein | |
| 78 | t4301 | 4463686..4464858 [-], 1173 | probable major tail sheath protein | |
| 79 | sopE | 4465393..4466115 [+], 723 | invasion-associated secreted protein | |
| 80 | t4304 | 4466313..4466720 [-], 408 | putative phage tail fiber protein | |
| 81 | t4305 | 4466727..4468346 [-], 1620 | probable phage tail fiber protein | |
| 82 | t4306 | 4468343..4468948 [-], 606 | putative phage tail protein | |
| 83 | t4307 | 4468941..4469849 [-], 909 | probable phage baseplate assembly protein | |
| 84 | t4308 | 4469836..4470195 [-], 360 | phage baseplate assembly protein | |
| 85 | t4309 | 4470192..4470770 [-], 579 | putative phage baseplate assembly protein | |
| 86 | t4310 | 4470839..4471285 [-], 447 | putative phage tail completion protein | |
| 87 | t4312 | 4471805..4472230 [-], 426 | putative regulatory protein | |
| 88 | t4313 | 4472230..4472607 [-], 378 | putative membrane protein | |
| 89 | nucD | 4472612..4473082 [-], 471 | putative lysozyme | |
| 90 | nucE | 4473102..4473317 [-], 216 | possible secretion protein | |
| 91 | t4316 | 4473321..4473524 [-], 204 | putative phage tail protein | |
| 92 | t4317 | 4473524..4473988 [-], 465 | putative capsid completion protein | |
| 93 | t4318 | 4474082..4474732 [-], 651 | putative phage terminase | |
| 94 | t4319 | 4474736..4475800 [-], 1065 | putative major capsid protein | |
| 95 | t4320 | 4475817..4476650 [-], 834 | putative capsid protein | |
| 96 | t4322 | 4476793..4478559 [+], 1767 | probable terminase subunit | |
| 97 | t4321 | 4478556..4479602 [+], 1047 | probable capsid portal protein | |
| 98 | t4323 | 4479651..4480346 [-], 696 | hypothetical protein | |
| 99 | t4324 | 4480366..4481430 [-], 1065 | hypothetical protein | |
| 100 | t4325 | 4481427..4482491 [-], 1065 | hypothetical protein | |
| 101 | t4326 | 4482501..4482719 [-], 219 | hypothetical protein | |
| 102 | t4328 | 4483416..4485824 [-], 2409 | conserved hypothetical protein | |
| 103 | t4329 | 4485815..4486675 [-], 861 | DNA adenine methylase | |
| 104 | t4330 | 4486672..4487256 [-], 585 | putative exonuclease | |
| 105 | t4331 | 4487253..4487480 [-], 228 | hypothetical protein | |
| 106 | t4332 | 4487480..4487713 [-], 234 | hypothetical protein | |
| 107 | t4333 | 4487781..4488122 [-], 342 | conserved hypothetical protein | |
| 108 | t4334 | 4488086..4488286 [-], 201 | hypothetical protein | |
| 109 | cII | 4488294..4488803 [-], 510 | phage regulatory protein | |
| 110 | apl | 4488836..4489078 [-], 243 | phage regulatory protein | |
| 111 | cI | 4489195..4489827 [+], 633 | phage repressor protein cI | |
| 112 | t4338 | 4489831..4490856 [+], 1026 | phage integrase | Integrase |
| 113 | t4340 | 4491584..4491697 [+], 114 | hypothetical protein | |
| 114 | t4341 | 4491933..4492220 [+], 288 | hypothetical protein | |
| 115 | t4342 | 4492231..4493121 [+], 891 | Putative methyltransferase | |
| 116 | yjhP | 4493121..4493867 [+], 747 | conserved hypothetical protein | |
| 117 | vexE | 4494169..4496139 [-], 1971 | Vi polysaccharide export protein | |
| 118 | vexD | 4496159..4497463 [-], 1305 | Vi polysaccharide export inner-membrane protein | |
| 119 | vexC | 4497486..4498181 [-], 696 | Vi polysaccharide export ATP-binding protein | |
| 120 | vexB | 4498207..4499001 [-], 795 | Vi polysaccharide export inner-membrane protein | |
| 121 | vexA | 4499011..4500078 [-], 1068 | Vi polysaccharide export protein | |
| 122 | tviE | 4500123..4501859 [-], 1737 | Vi polysaccharide biosynthesis protein TviE | |
| 123 | tviD | 4501859..4504354 [-], 2496 | Vi polysaccharide biosynthesis protein | |
| 124 | tviC | 4504378..4505424 [-], 1047 | Vi polysaccharide biosynthesis protein, epimerase | |
| 125 | tviB | 4505427..4506704 [-], 1278 | Vi polysaccharide biosynthesis protein, UDP-glucose/GDP-mannose dehydrogenase | |
| 126 | tviA | 4506949..4507488 [-], 540 | Vi polysaccharide biosynthesis protein | |
| 127 | t4354 | 4508029..4508217 [+], 189 | hypothetical protein | |
| 128 | t4355 | 4508342..4509853 [+], 1512 | putative DNA helicase | TraI_F, T4SS component |
| 129 | t4356 | 4509837..4511426 [+], 1590 | hypothetical protein | Relaxase, MOBH Family |
| 130 | t4357 | 4511590..4512603 [+], 1014 | probable phage integrase | |
| 131 | t4358 | 4513030..4513323 [+], 294 | hypothetical protein | |
| 132 | t4359 | 4513320..4513808 [+], 489 | conserved hypothetical protein | |
| 133 | t4360 | 4513988..4514440 [-], 453 | putative membrane protein | |
| 134 | t4361 | 4515077..4517362 [-], 2286 | hypothetical protein | |
| 135 | t4362 | 4517443..4519116 [-], 1674 | putative membrane protein | |
| 136 | t4363 | 4519663..4520124 [+], 462 | hypothetical protein | |
| 137 | cII | 4520121..4520342 [+], 222 | transcriptional regulatory protein | |
| 138 | t4365 | 4520376..4520519 [-], 144 | hypothetical protein | |
| 139 | t4366 | 4520735..4521061 [-], 327 | hypothetical protein | |
| 140 | t4367 | 4521199..4521981 [+], 783 | hypothetical protein | |
| 141 | t4368 | 4522006..4522218 [-], 213 | putative membrane protein | |
| 142 | - | 4522962..4524245 [-], 1284 | putative membrane protein | |
| 143 | int | 4524556..4525815 [-], 1260 | bacteriophage integrase | |
| 144 | yjdC | 4526166..4526741 [-], 576 | putative transcriptional regulator | |
| 145 | dsbD | 4526778..4528481 [-], 1704 | thiol:disulfide interchange protein DsbD | |
| 146 | cutA | 4528457..4528804 [-], 348 | periplasmic divalent cation tolerance protein CutA | |
| 147 | dcuA | 4528925..4530226 [-], 1302 | anaerobic C4-dicarboxylate transporter | |