The graph information of ICESa2 components from AM990992 |
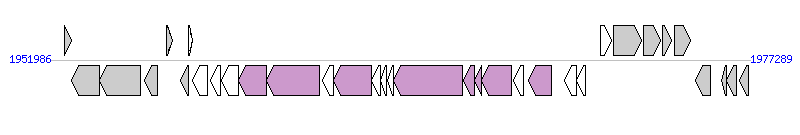 |
| Complete gene list of ICESa2 from AM990992 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | SAPIG1843 | 1952509..1952766 [+], 258 | conserved hypothetical protein | |
| 2 | menC | 1952763..1953764 [-], 1002 | O-succinylbenzoic acid (OSB) synthetase | |
| 3 | menE | 1953769..1955247 [-], 1479 | O-succinylbenzoate-CoA ligase | |
| 4 | SAPIG1845 | 1955406..1955888 [-], 483 | lipoprotein, putative | |
| 5 | SAPIG1846 | 1956200..1956427 [+], 228 | hypothetical protein | |
| 6 | SAPIG1847 | 1956685..1956984 [-], 300 | transposase InsI for insertion sequence element IS30B/C/D | |
| 7 | SAPIG1848 | 1956986..1957126 [+], 141 | hypothetical protein | |
| 8 | SAPIG1849 | 1957139..1957669 [-], 531 | transposase of | |
| 9 | SAPIG1850 | 1957781..1958143 [-], 363 | lipoprotein, putative | |
| 10 | SAPIG1851 | 1958190..1958783 [-], 594 | conserved hypothetical protein | |
| 11 | SAPIG1852 | 1958789..1959817 [-], 1029 | transfer complex protein TraG | Orf13_p, T4SS component |
| 12 | SAPIG1853 | 1959807..1961738 [-], 1932 | membrane protein, putative | YddG, T4SS component |
| 13 | SAPIG1854 | 1961838..1962239 [-], 402 | hypothetical protein | |
| 14 | SAPIG1855 | 1962244..1963602 [-], 1359 | ftsk/spoiiie family protein | YdcQ, T4SS component |
| 15 | SAPIG1856 | 1963607..1963939 [-], 333 | hypothetical protein | |
| 16 | SAPIG1857 | 1963936..1964166 [-], 231 | hypothetical protein | |
| 17 | SAPIG1858 | 1964170..1964388 [-], 219 | hypothetical protein | |
| 18 | SAPIG1859 | 1964400..1966895 [-], 2496 | conserved hypothetical protein | YddE, T4SS component |
| 19 | SAPIG1860 | 1966930..1967319 [-], 390 | conserved hypothetical protein | YddD, T4SS component |
| 20 | SAPIG1861 | 1967325..1967585 [-], 261 | conserved hypothetical protein | YddC, T4SS component |
| 21 | SAPIG1862 | 1967590..1968666 [-], 1077 | conserved hypothetical protein | YddB, T4SS component |
| 22 | SAPIG1863 | 1968741..1969103 [-], 363 | conserved hypothetical protein | |
| 23 | SAPIG1864 | 1969283..1970113 [-], 831 | replication initiation factor family protein | Relaxase, MOBT Family |
| 24 | SAPIG1865 | 1970588..1971013 [-], 426 | hypothetical protein | |
| 25 | SAPIG1866 | 1971035..1971358 [-], 324 | hypothetical protein | |
| 26 | SAPIG1867 | 1971879..1972289 [+], 411 | excalibur domain protein | |
| 27 | SAPIG1868 | 1972370..1973365 [+], 996 | conserved hypothetical protein | |
| 28 | SAPIG1869 | 1973441..1974067 [+], 627 | lipoprotein, putative | |
| 29 | SAPIG1870 | 1974108..1974452 [+], 345 | conserved hypothetical protein | |
| 30 | SAPIG1871 | 1974550..1975122 [+], 573 | conserved hypothetical protein | |
| 31 | SAPIG1872 | 1975320..1975877 [-], 558 | conserved hypothetical protein | |
| 32 | SAPIG1873 | 1976248..1976424 [-], 177 | conserved hypothetical protein | |
| 33 | SAPIG1874 | 1976435..1976818 [-], 384 | conserved hypothetical protein | |
| 34 | SAPIG1875 | 1976920..1977225 [-], 306 | transposase | |
