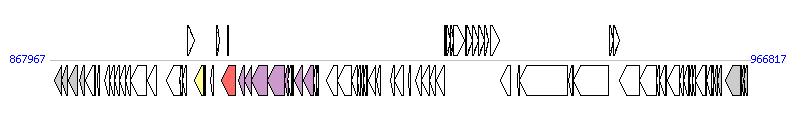The graph information of ICESsu05ZYH33-1 components from CP000407 |
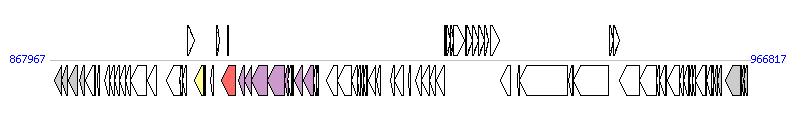 |
| Complete gene list of ICESsu05ZYH33-1 from CP000407 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | SSU05_0899 | 868566..869609 [-], 1044 | hypothetical protein | |
| 2 | SSU05_0900 | 869635..870474 [-], 840 | Uncharacterized protein conserved in bacteria | |
| 3 | SSU05_0901 | 870548..871882 [-], 1335 | NAD(FAD)-utilizing enzyme possibly involved in translation | |
| 4 | SSU05_0902 | 872078..872827 [-], 750 | Predicted hydrolase (HAD superfamily) | |
| 5 | SSU05_0903 | 873012..874223 [-], 1212 | site-specific recombinase, phage integrase family | |
| 6 | SSU05_0904 | 874234..874461 [-], 228 | hypothetical protein | |
| 7 | SSU05_0905 | 874740..874961 [-], 222 | transcriptional regulator, Cro/CI family | |
| 8 | SSU05_0906 | 875685..876392 [-], 708 | NisK | |
| 9 | SSU05_0907 | 876377..877060 [-], 684 | NisR | |
| 10 | SSU05_0908 | 877091..877744 [-], 654 | hypothetical protein | |
| 11 | SSU05_0909 | 877796..878593 [-], 798 | hypothetical protein | |
| 12 | SSU05_0910 | 878577..879293 [-], 717 | putative transport ATP-binding protein | |
| 13 | SSU05_0911 | 879314..881536 [-], 2223 | ABC transporter, NBP/MSD fusion protein | |
| 14 | SSU05_0912 | 881803..882948 [-], 1146 | putative asparagine synthetase | |
| 15 | SSU05_0913 | 884460..886325 [-], 1866 | Chromosome segregation ATPase | |
| 16 | SSU05_0914 | 886312..886677 [-], 366 | unknown protein | |
| 17 | SSU05_0915 | 886677..887252 [-], 576 | hypothetical protein | |
| 18 | SSU05_0916 | 887407..888303 [+], 897 | putative Abi-alpha protein | |
| 19 | SSU05_0917 | 888425..889642 [-], 1218 | Tn916, transposase | Integrase |
| 20 | SSU05_0918 | 889724..889927 [-], 204 | hypothetical protein | |
| 21 | SSU05_0919 | 890615..891046 [-], 432 | hypothetical protein | |
| 22 | SSU05_0920 | 891541..891894 [+], 354 | Tn916, transcriptional regulator, putative | |
| 23 | SSU05_0922 | 892240..894174 [-], 1935 | Translation elongation factor (GTPases) | AR |
| 24 | SSU05_0921 | 893067..893219 [+], 153 | hypothetical protein | |
| 25 | SSU05_0923 | 894536..895468 [-], 933 | Tn916, hypothetical protein | Orf13_Tn, T4SS component |
| 26 | SSU05_0924 | 895465..896466 [-], 1002 | hypothetical protein | Orf14_Tn, T4SS component |
| 27 | SSU05_0925 | 896463..898640 [-], 2178 | membrane protein, putative | Orf15_Tn, T4SS component |
| 28 | SSU05_0926 | 898643..901099 [-], 2457 | hypothetical protein | Orf16_Tn, T4SS component |
| 29 | SSU05_0927 | 901074..901580 [-], 507 | hypothetical protein | Orf17_Tn, T4SS component |
| 30 | SSU05_0928 | 901555..902052 [-], 498 | hypothetical protein | |
| 31 | SSU05_0929 | 902169..902390 [-], 222 | hypothetical protein | Orf19_Tn, T4SS component |
| 32 | SSU05_0930 | 902433..903638 [-], 1206 | Tn916, transcriptional regulator, putative | Relaxase, MOBT Family |
| 33 | SSU05_0931 | 903816..905201 [-], 1386 | hypothetical protein | Orf21_Tn, T4SS component |
| 34 | SSU05_0932 | 905230..905616 [-], 387 | hypothetical protein | |
| 35 | SSU05_0933 | 905632..905946 [-], 315 | hypothetical protein | |
| 36 | SSU05_0934 | 907006..908550 [-], 1545 | DNA helicase | |
| 37 | SSU05_0935 | 908507..910594 [-], 2088 | hypothetical protein | |
| 38 | SSU05_0936 | 910591..911367 [-], 777 | Signal recognition particle GTPase | |
| 39 | SSU05_0937 | 911367..911873 [-], 507 | Predicted transcriptional regulator | |
| 40 | SSU05_0938 | 911913..912266 [-], 354 | ATPases with chaperone activity, ATP-binding subunit | |
| 41 | SSU05_0939 | 912253..912642 [-], 390 | Methyl-accepting chemotaxis protein | |
| 42 | SSU05_0940 | 912639..912866 [-], 228 | hypothetical protein | |
| 43 | SSU05_0941 | 912920..913996 [-], 1077 | DNA primase (type) | |
| 44 | SSU05_0942 | 914035..914673 [-], 639 | hypothetical protein | |
| 45 | SSU05_0943 | 916074..916673 [-], 600 | DNA-binding response regulator, putative | |
| 46 | SSU05_0944 | 916651..917841 [-], 1191 | possible two-component sensor histidine kinase | |
| 47 | SSU05_0945 | 918568..918852 [-], 285 | hypothetical protein | |
| 48 | SSU05_0946 | 919557..920471 [-], 915 | cdd4 | |
| 49 | SSU05_0947 | 920443..921546 [-], 1104 | ABC-type multidrug transport system, ATPase and permease component | |
| 50 | SSU05_0948 | 921488..922372 [-], 885 | cytolysin B transport protein | |
| 51 | SSU05_0949 | 922525..923637 [-], 1113 | lantibiotic modifying enzyme | |
| 52 | SSU05_0950 | 923737..923856 [+], 120 | hypothetical protein | |
| 53 | SSU05_0951 | 923964..924461 [+], 498 | DNA recombinase, putative | |
| 54 | SSU05_0952 | 924462..924878 [+], 417 | Recombinase | |
| 55 | SSU05_0953 | 924880..926442 [+], 1563 | DNA recombinase, putative | |
| 56 | SSU05_0954 | 926585..926809 [+], 225 | hypothetical protein | |
| 57 | SSU05_0955 | 926824..927693 [+], 870 | hypothetical protein | |
| 58 | SSU05_0956 | 927674..928408 [+], 735 | hypothetical protein | |
| 59 | SSU05_0957 | 928441..929304 [+], 864 | putative aminoglycoside 6-adenylyltansferase | |
| 60 | SSU05_0958 | 929348..929875 [+], 528 | putative adenine phosphoribosyltransferase | |
| 61 | SSU05_0959 | 930103..931422 [+], 1320 | putative transposase | |
| 62 | SSU05_0960 | 931611..933008 [-], 1398 | SalB | |
| 63 | SSU05_0961 | 933916..934215 [-], 300 | hypothetical protein | |
| 64 | SSU05_0962 | 934286..941110 [-], 6825 | SNF2 family protein | |
| 65 | SSU05_0963 | 941160..941714 [-], 555 | hypothetical protein | |
| 66 | SSU05_0964 | 941695..941886 [-], 192 | hypothetical protein | |
| 67 | SSU05_0965 | 941887..946839 [-], 4953 | agglutinin receptor | |
| 68 | SSU05_0966 | 946954..947544 [+], 591 | Predicted transcriptional regulator | |
| 69 | SSU05_0967 | 947541..948386 [+], 846 | Uncharacterized conserved protein | |
| 70 | SSU05_0968 | 948449..951250 [-], 2802 | Tn5252, Orf28 | |
| 71 | SSU05_0969 | 951252..953603 [-], 2352 | Type IV secretory pathway, VirB4 component | |
| 72 | SSU05_0970 | 953560..954015 [-], 456 | hypothetical protein | |
| 73 | SSU05_0971 | 953975..954829 [-], 855 | ABC-type cobalt transport system, permease component CbiQ and related transporters | |
| 74 | SSU05_0972 | 954848..955069 [-], 222 | hypothetical protein | |
| 75 | SSU05_0973 | 955109..956926 [-], 1818 | Type IV secretory pathway, VirD4 component | |
| 76 | SSU05_0974 | 956926..957495 [-], 570 | hypothetical protein | |
| 77 | SSU05_0975 | 957492..958082 [-], 591 | protease, putative | |
| 78 | SSU05_0976 | 958079..958318 [-], 240 | hypothetical protein | |
| 79 | SSU05_0977 | 958321..958710 [-], 390 | Arsenate reductase and related proteins, glutaredoxin family | |
| 80 | SSU05_0978 | 958715..959143 [-], 429 | hypothetical protein | |
| 81 | SSU05_0979 | 959127..960482 [-], 1356 | C-5 cytosine-specific DNA methylase | |
| 82 | SSU05_0980 | 960534..960638 [-], 105 | hypothetical protein | |
| 83 | SSU05_0981 | 960631..961449 [-], 819 | hypothetical protein | |
| 84 | SSU05_0982 | 961446..961631 [-], 186 | hypothetical protein | |
| 85 | SSU05_0983 | 961818..962183 [-], 366 | Ribosomal protein L7/L12 | |
| 86 | SSU05_0984 | 962249..962779 [-], 531 | Ribosomal protein L10 | |
| 87 | SSU05_0985 | 963320..965461 [-], 2142 | DNA topoisomerase I | |
| 88 | SSU05_0986 | 965515..965652 [-], 138 | Predicted Rossmann fold nucleotide-binding protein involved in DNA uptake | |
| 89 | SSU05_0987 | 965671..965970 [-], 300 | Predicted Rossmann fold nucleotide-binding protein involved in DNA uptake | |
| 90 | SSU05_0988 | 965967..966443 [-], 477 | Predicted Rossmann fold nucleotide-binding protein involved in DNA uptake | |