The graph information of ICEA(PG2) components from CU179680 |
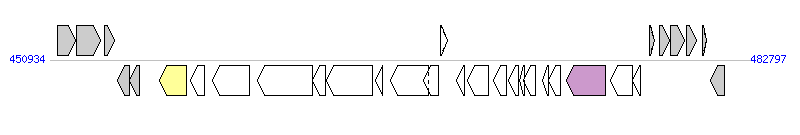 |
| Complete gene list of ICEA(PG2) from CU179680 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | MAG3810 | 451278..452084 [+], 807 | Hypothetical protein | |
| 2 | MAG3820 | 452130..453233 [+], 1104 | GTP binding protein | |
| 3 | MAG3830 | 453422..453880 [+], 459 | Hypothetical protein | |
| 4 | hpt | 453996..454544 [-], 549 | Hypoxanthine guanine phosphoribosyltransferase(HGPRT) | |
| 5 | greA | 454534..455025 [-], 492 | Transcription elongation factor greA | |
| 6 | MAG3860 | 455934..457166 [-], 1233 | Conserved hypothetical protein | Integrase |
| 7 | MAG3870 | 457338..457976 [-], 639 | Conserved hypothetical protein | |
| 8 | MAG3880 | 458312..460006 [-], 1695 | Pseudogen of conserved hypothetical protein (C terminal part) | |
| 9 | MAG3890 | 460357..462870 [-], 2514 | Pseudogen of conserved hypothetical protein (N terminal part) | |
| 10 | MAG3900 | 462873..463460 [-], 588 | Conserved hypothetical protein | |
| 11 | MAG3910 | 463503..465608 [-], 2106 | Pseudogene of TraE/TrsE family NTPase(C terminal part) | |
| 12 | MAG3920 | 465767..466087 [-], 321 | Pseudogene of TraE/TrsE family NTPase(N terminal part) | |
| 13 | MAG3930 | 466414..468168 [-], 1755 | Pseudogen of conserved hypothetical protein (C terminal part) | |
| 14 | MAG3940 | 467933..468604 [-], 672 | Pseudogen of conserved hypothetical protein (N terminal part) | |
| 15 | MAG3950 | 468730..469005 [+], 276 | Hypothetical protein | |
| 16 | MAG3960 | 469428..469802 [-], 375 | Conserved hypothetical protein, truncated in N terminal | |
| 17 | MAG3970 | 469937..470911 [-], 975 | Conserved hypothetical protein, predicted lipoprotein, truncated in C ter | |
| 18 | MAG3980 | 471119..471697 [-], 579 | Conserved hypothetical protein | |
| 19 | MAG3990 | 471807..472244 [-], 438 | Conserved hypothetical protein | |
| 20 | MAG4000 | 472282..472524 [-], 243 | Hypothetical protein | |
| 21 | MAG4010 | 472505..473020 [-], 516 | Hypothetical protein | |
| 22 | MAG4020 | 473350..473616 [-], 267 | Hypothetical protein | |
| 23 | MAG4030 | 473623..474183 [-], 561 | Conserved hypothetical protein | |
| 24 | MAG4040 | 474429..476201 [-], 1773 | Conserved hypothetical protein | T4CP |
| 25 | MAG4050 | 476439..477428 [-], 990 | Conserved hypothetical protein | |
| 26 | MAG4060 | 477432..477797 [-], 366 | Conserved hypothetical protein, truncated in C terminal | |
| 27 | MAG4070 | 478206..478472 [+], 267 | Hypothetical protein | |
| 28 | MAG4080 | 478695..479150 [+], 456 | Hypothetical protein | |
| 29 | MAG4090 | 479172..479804 [+], 633 | Hypothetical protein | |
| 30 | MAG4100 | 479917..480354 [+], 438 | Pseudogene of Transposase (N terminal part) | |
| 31 | MAG4110 | 480625..480819 [+], 195 | Pseudogene of Transposase (C terminal part) | |
| 32 | MAG4120 | 480978..481616 [-], 639 | Oxidoreductase, potentially truncated inN terminal | |
