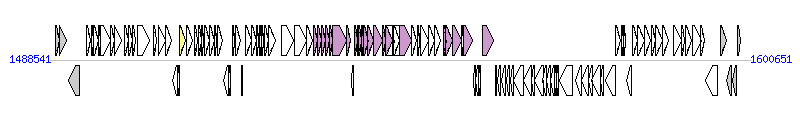The graph information of ICE-GI3 components from AM902716 |
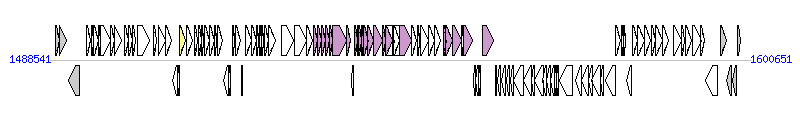 |
| Complete gene list of ICE-GI3 from AM902716 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | Bpet1435 | 1489427..1490008 [+], 582 | transcriptional regulator, TetR-family | |
| 2 | Bpet1436 | 1490093..1491262 [+], 1170 | putative Transmembrane efflux protein of the MFS type | |
| 3 | intB13 | 1491479..1493314 [-], 1836 | phage-related integrase | |
| 4 | Bpet1438 | 1494345..1495100 [+], 756 | conserved hypothetical protein | |
| 5 | alpA3 | 1495220..1495432 [+], 213 | predicted transcriptional regulator (phage-related) | |
| 6 | parA3 | 1495476..1496351 [+], 876 | hypothetical protein | |
| 7 | Bpet1441 | 1496335..1496592 [+], 258 | conserved hypothetical protein | |
| 8 | Bpet1442 | 1496585..1498237 [+], 1653 | conserved hypothetical protein | |
| 9 | Bpet1443 | 1498253..1498813 [+], 561 | conserved hypothetical protein | |
| 10 | Bpet1444 | 1498817..1500061 [+], 1245 | conserved hypothetical protein | |
| 11 | Bpet1445 | 1500392..1501171 [+], 780 | conserved hypothetical protein | |
| 12 | Bpet1446 | 1501168..1501695 [+], 528 | conserved hypothetical protein | |
| 13 | ssb3 | 1501769..1502209 [+], 441 | - | |
| 14 | topB3 | 1502488..1504500 [+], 2013 | DNA topoisomerase III | |
| 15 | Bpet1449 | 1505037..1505642 [+], 606 | putative DNA-cytosine methyltransferase | |
| 16 | Bpet1450 | 1505848..1506987 [+], 1140 | integrase/recombinase | |
| 17 | Bpet1451 | 1506987..1507982 [+], 996 | integrase/recombinase | |
| 18 | Bpet1452 | 1508109..1508999 [-], 891 | probable transposase | |
| 19 | tnp8 | 1508996..1509283 [-], 288 | transposase | |
| 20 | int1 | 1509356..1510309 [+], 954 | putative integrase/recombinase | Integrase |
| 21 | Bpet1455 | 1510428..1511417 [+], 990 | DNA-cytosine methyltransferase | |
| 22 | Bpet1456 | 1511662..1512219 [+], 558 | conserved hypothetical protein | |
| 23 | Bpet1457 | 1512376..1512762 [+], 387 | conserved hypothetical protein | |
| 24 | Bpet1458 | 1512784..1513176 [+], 393 | conserved hypothetical protein | |
| 25 | Bpet1459 | 1513349..1514035 [+], 687 | conserved hypothetical protein | |
| 26 | Bpet1460 | 1514115..1514852 [+], 738 | conserved hypothetical protein | |
| 27 | Bpet1461 | 1514950..1515228 [+], 279 | conserved hypothetical protein | |
| 28 | Bpet1462 | 1515302..1516243 [+], 942 | conserved hypothetical protein | |
| 29 | Bpet1463 | 1516254..1517144 [-], 891 | probable transposase | |
| 30 | tnp9 | 1517141..1517428 [-], 288 | transposase | |
| 31 | Bpet1465 | 1517756..1518109 [+], 354 | conserved hypothetical protein | |
| 32 | Bpet1466 | 1518255..1519097 [+], 843 | conserved hypothetical protein | |
| 33 | Bpet1467 | 1519151..1519294 [-], 144 | Potential Pseudomonas putida clc transposon | |
| 34 | Bpet1468 | 1519875..1520792 [+], 918 | hypothetical protein | |
| 35 | Bpet1469 | 1520850..1521539 [+], 690 | conserved hypothetical protein | |
| 36 | Bpet1470 | 1521635..1521967 [+], 333 | conserved hypothetical protein | |
| 37 | Bpet1471 | 1522064..1522471 [+], 408 | - | |
| 38 | Bpet1472 | 1522488..1522748 [+], 261 | hypothetical protein predicted by Glimmer/Critica | |
| 39 | Bpet1473 | 1522825..1523475 [+], 651 | conserved hypothetical protein | |
| 40 | Bpet1474 | 1523540..1524649 [+], 1110 | conserved hypothetical protein | |
| 41 | ltrA2 | 1525670..1527490 [+], 1821 | Mobile mitochondrial group II intron of COX1 which is involved in pre-mRNA splicing and in deletion of introns from mitochondrial DNA | |
| 42 | Bpet1476 | 1527718..1529565 [+], 1848 | conserved plasmid related protein | |
| 43 | Bpet1477 | 1529679..1530548 [+], 870 | hypothetical protein predicted by Glimmer/Critica | |
| 44 | Bpet1478 | 1530709..1531308 [+], 600 | putative secreted protein | Tfc2, T4SS component |
| 45 | Bpet1479 | 1531305..1531955 [+], 651 | putative membrane protein | Tfc2, T4SS component |
| 46 | Bpet1480 | 1531970..1532689 [+], 720 | conserved protein, putatively exported | Tfc3, T4SS component |
| 47 | Bpet1481 | 1532671..1533261 [+], 591 | conserved exported protein | Tfc4, T4SS component |
| 48 | Bpet1482 | 1533258..1533806 [+], 549 | conserved hypothetical protein | Tfc5, T4SS component |
| 49 | Bpet1483 | 1533811..1535997 [+], 2187 | putative plasmid-transfer-protein | Tfc6, T4SS component |
| 50 | Bpet1484 | 1535994..1536743 [+], 750 | putative Membrane protein | Tfc8, T4SS component |
| 51 | Bpet1485 | 1536780..1537184 [-], 405 | conserved hypothetical protein | |
| 52 | Bpet1486 | 1537370..1537753 [+], 384 | putative membrane protein | Tfc9, T4SS component |
| 53 | Bpet1487 | 1537750..1537983 [+], 234 | conserved hypothetical protein | |
| 54 | Bpet1488 | 1537994..1538359 [+], 366 | conserved hypothetical protein | Tfc10, T4SS component |
| 55 | Bpet1489 | 1538372..1538782 [+], 411 | putative membrane protein | Tfc11, T4SS component |
| 56 | Bpet1490 | 1538779..1539471 [+], 693 | conserved hypothetical protein | Tfc12, T4SS component |
| 57 | Bpet1491 | 1539468..1540400 [+], 933 | putative secreted protein | Tfc13, T4SS component |
| 58 | Bpet1492 | 1540390..1541808 [+], 1419 | putative exported protein | Tfc14, T4SS component |
| 59 | Bpet1493 | 1541789..1542229 [+], 441 | putative lipoprotein | Tfc15, T4SS component |
| 60 | bp050705_PS_02 | 1542229..1546381 [+], 4153 | - | |
| 61 | Bpetpseudo_04 | 1542229..1543317 [+], 1089 | pseudogene | Tfc16, T4SS component |
| 62 | tnp10 | 1543350..1543637 [+], 288 | transposase | |
| 63 | Bpet1495 | 1543634..1544524 [+], 891 | probable transposase | |
| 64 | Bpetpseudo_05 | 1544570..1546381 [+], 1812 | pseudogene | Tfc16, T4SS component |
| 65 | Bpet1496 | 1546395..1547159 [+], 765 | putative protein-disulfide isomerase | |
| 66 | Bpet1497 | 1547343..1547789 [+], 447 | putative DNA repair protein RadC | |
| 67 | Bpet1498 | 1547845..1549104 [+], 1260 | putative integrase/recombinase | |
| 68 | Bpet1499 | 1549101..1550093 [+], 993 | putative integrase/recombinase | |
| 69 | Bpet1500 | 1550090..1551100 [+], 1011 | putative integrase/recombinase | |
| 70 | Bpet1501 | 1551600..1552046 [+], 447 | conserved hypothetical protein | Tfc24, T4SS component |
| 71 | Bpet1502 | 1551923..1552990 [+], 1068 | conserved hypothetical protein | Tfc23, T4SS component |
| 72 | Bpet1503 | 1553000..1554397 [+], 1398 | conserved hypothetical protein | Tfc22, T4SS component |
| 73 | Bpet1504 | 1554394..1554753 [+], 360 | hypothetical protein predicted by Glimmer/Critica | |
| 74 | Bpet1505 | 1554769..1556286 [+], 1518 | conserved hypothetical protein | Tfc19, T4SS component |
| 75 | Bpet1506 | 1556300..1556677 [-], 378 | putative transposon | |
| 76 | Bpet1507 | 1556705..1557163 [-], 459 | conserved hypothetical protein | |
| 77 | Bpet1508 | 1557163..1557480 [-], 318 | probable suppressor protein | |
| 78 | Bpet1509 | 1557786..1559633 [+], 1848 | putative helicase | Relaxase, MOBH Family |
| 79 | Bpet1510 | 1559933..1560112 [-], 180 | putative transposon | |
| 80 | Bpet1511 | 1560148..1560621 [-], 474 | putative transcriptional regulator | |
| 81 | Bpet1512 | 1560803..1561570 [-], 768 | similar to glucose 1-dehydrogenase | |
| 82 | Bpet1513 | 1561652..1562170 [-], 519 | Biphenyl dioxygenase small subunit | |
| 83 | Bpetpseudo_06 | 1562213..1562752 [-], 540 | pseudogene | |
| 84 | Bpet1514 | 1562792..1564339 [-], 1548 | putative transposase | |
| 85 | Bpet1515 | 1564448..1565650 [-], 1203 | probable dihydrolipoamide dehydrogenase | |
| 86 | Bpet1516 | 1565629..1566108 [-], 480 | Putative acyl-CoA thioester hydrolase | |
| 87 | Bpet1517 | 1566127..1567602 [-], 1476 | probable glutathione reductase | |
| 88 | Bpet1518 | 1567491..1568060 [-], 570 | probable glutathione S-transferase | |
| 89 | gst2 | 1568129..1568749 [-], 621 | putative glutathione transferase | |
| 90 | Bpet1520 | 1568763..1569353 [-], 591 | putative enzyme | |
| 91 | Bpet1521 | 1569368..1569670 [-], 303 | hypothetical protein predicted by Glimmer/Critica | |
| 92 | Bpet1522 | 1569683..1569988 [-], 306 | conserved hypothetical protein | |
| 93 | Bpet1523 | 1570186..1572237 [-], 2052 | transposition-related fusion-protein | |
| 94 | Bpet1524 | 1572677..1573705 [-], 1029 | glycosyltransferase | |
| 95 | Bpet1525 | 1573702..1574760 [-], 1059 | glycosyltransferase | |
| 96 | Bpet1526 | 1574753..1575355 [-], 603 | acetyltransferase | |
| 97 | Bpet1527 | 1575339..1576721 [-], 1383 | conserved hypothetical protein | |
| 98 | Bpet1528 | 1576782..1577165 [-], 384 | putative transposase | |
| 99 | Bpet1529 | 1577568..1579115 [-], 1548 | putative transposase | |
| 100 | Bpet1530 | 1579174..1579845 [+], 672 | ISSod11, transposase | |
| 101 | Bpet1531 | 1580078..1580320 [+], 243 | conserved hypothetical protein | |
| 102 | Bpet1532 | 1580338..1580688 [+], 351 | conserved hypothetical protein | |
| 103 | clcR | 1580832..1581716 [-], 885 | transcriptional regulator clcR | |
| 104 | clcA | 1581886..1582668 [+], 783 | catechol 1,2-dioxygenase | |
| 105 | catB2 | 1582665..1583777 [+], 1113 | muconate cycloisomerase I | |
| 106 | bug36 | 1583804..1584787 [+], 984 | putative secreted protein | |
| 107 | Bpet1537 | 1584809..1585519 [+], 711 | probable carboxymethylenebutenolidase | |
| 108 | Bpet1538 | 1585516..1586574 [+], 1059 | maleylacetate reductase | |
| 109 | Bpet1539 | 1586690..1587673 [+], 984 | transcriptional regulator, AraC family | |
| 110 | Bpet1540 | 1588469..1589731 [+], 1263 | probable Ring hydroxylating alpha subunit | |
| 111 | Bpet1541 | 1589734..1590213 [+], 480 | conserved hypothetical protein | |
| 112 | Bpet1542 | 1590311..1591288 [+], 978 | Vanillate O-demethylase oxidoreductase | |
| 113 | Bpet1543 | 1591493..1592878 [+], 1386 | benzoate transport protein, putative | |
| 114 | Bpet1544 | 1592784..1593284 [+], 501 | hypothetical protein predicted by Glimmer/Critica | |
| 115 | intB13 | 1593493..1595466 [-], 1974 | phage-related integrase | |
| 116 | murB | 1595859..1596920 [+], 1062 | UDP-N-acetylmuramate dehydrogenase | |
| 117 | dapB | 1596967..1597758 [-], 792 | - | |
| 118 | omlA | 1597819..1598547 [-], 729 | outer membrane lipoprotein OmlA | |
| 119 | fur | 1598708..1599130 [+], 423 | ferric uptake regulation protein | |