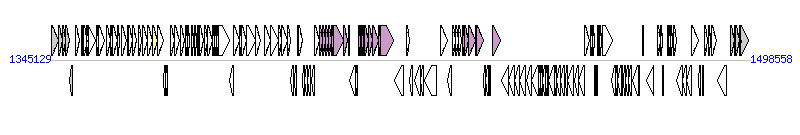The graph information of ICE-GI2 components from AM902716 |
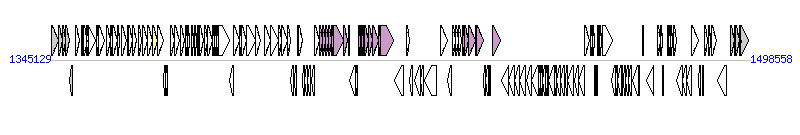 |
| Complete gene list of ICE-GI2 from AM902716 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | Bpet1283 | 1345519..1347024 [+], 1506 | putative long-chain-fatty-acid--CoA ligase | |
| 2 | Bpet1284 | 1347021..1347806 [+], 786 | putative Citrate synthase | |
| 3 | Bpet1285 | 1347809..1348561 [+], 753 | putative oxidoreductase | |
| 4 | Bpet1286 | 1348522..1349400 [+], 879 | probable short chain dehydrogenase | |
| 5 | Bpet1287 | 1349423..1350037 [-], 615 | - | |
| 6 | Bpet1288 | 1350756..1351655 [+], 900 | transcriptional regulator, LysR-family | |
| 7 | Bpet1289 | 1352135..1352347 [+], 213 | putative transcriptional regulator (phage-related) | |
| 8 | parA2 | 1352385..1353248 [+], 864 | conserved hypothetical protein | |
| 9 | ig_0609 | 1353232..1353489 [+], 258 | conserved hypothetical protein | |
| 10 | Bpet1291 | 1353482..1355206 [+], 1725 | conserved hypothetical protein | |
| 11 | Bpet1292 | 1355218..1355778 [+], 561 | conserved hypothetical protein | |
| 12 | Bpet1293 | 1355781..1356971 [+], 1191 | hypothetical protein | |
| 13 | Bpet1294 | 1357574..1358395 [+], 822 | conserved hypothetical protein | |
| 14 | Bpet1295 | 1358392..1358922 [+], 531 | probable transcriptional regulator | |
| 15 | Bpet1296 | 1358919..1359878 [+], 960 | adenine DNA methyltransferase protein | |
| 16 | ssb2 | 1359923..1360366 [+], 444 | - | |
| 17 | top3A | 1360772..1361959 [+], 1188 | pseudogene | |
| 18 | tnp5 | 1362023..1362310 [+], 288 | transposase | |
| 19 | Bpet1300 | 1362307..1363197 [+], 891 | probable transposase | |
| 20 | top3B | 1363278..1364081 [+], 804 | pseudogene | |
| 21 | Bpet1302 | 1364635..1365204 [+], 570 | putative C-5 cytosine-specific DNA methylase | |
| 22 | Bpet1303 | 1365410..1366549 [+], 1140 | integrase/recombinase | |
| 23 | Bpet1304 | 1366549..1367544 [+], 996 | integrase/recombinase | |
| 24 | Bpet1305 | 1367537..1368586 [+], 1050 | putative integrase/recombinase | Integrase |
| 25 | Bpet1306 | 1368705..1369904 [+], 1200 | DNA-cytosine methyltransferase | |
| 26 | Bpet1307 | 1370007..1370522 [-], 516 | probable acetyltransferase | |
| 27 | Bpet1308 | 1370522..1370800 [-], 279 | conserved hypothetical protein | |
| 28 | Bpet1309 | 1371604..1372317 [+], 714 | hypothetical protein predicted by Glimmer/Critica | |
| 29 | Bpet1310 | 1372475..1373446 [+], 972 | hypothetical protein predicted by Glimmer/Critica | |
| 30 | Bpet1311 | 1373700..1374512 [+], 813 | conserved hypothetical protein | |
| 31 | Bpet1312 | 1374834..1375199 [+], 366 | conserved hypothetical protein | |
| 32 | Bpet1313 | 1375305..1375646 [+], 342 | conserved plasmid protein | |
| 33 | Bpet1314 | 1376054..1376602 [+], 549 | hypothetical protein | |
| 34 | Bpet1315 | 1376709..1377317 [+], 609 | hypothetical protein | |
| 35 | Bpet1316 | 1377395..1377736 [+], 342 | hypothetical protein predicted by Glimmer/Critica | |
| 36 | Bpet1317 | 1377971..1378798 [+], 828 | Hypothetical protein (o273) | |
| 37 | Bpet1318 | 1378936..1379586 [+], 651 | conserved hypothetical protein | |
| 38 | Bpet1319 | 1379652..1380758 [+], 1107 | hypothetical protein predicted by Glimmer/Critica | |
| 39 | Bpet1320 | 1380838..1381164 [+], 327 | hypothetical protein predicted by Glimmer/Critica | |
| 40 | Bpet1321 | 1381254..1381559 [+], 306 | hypothetical protein predicted by Glimmer/Critica | |
| 41 | Bpet1322 | 1381685..1381996 [+], 312 | hypothetical protein | |
| 42 | Bpet1323 | 1382099..1384372 [+], 2274 | conserved plasmid related protein | |
| 43 | Bpet1324 | 1384388..1385263 [-], 876 | putative DNA-binding protein | |
| 44 | Bpet1325 | 1385419..1386660 [+], 1242 | 5-aminolevulinic acid synthase | |
| 45 | Bpet1326 | 1386661..1387161 [+], 501 | probable acetyltransferase | |
| 46 | Bpet1327 | 1387158..1388225 [+], 1068 | putative oxidoreductase | |
| 47 | asnB | 1388255..1390108 [+], 1854 | asparagine synthetase, glutamine-hydrolyzing | |
| 48 | Bpet1329 | 1390130..1391332 [+], 1203 | MFS transporter | |
| 49 | Bpet1330 | 1392232..1393371 [+], 1140 | putative monooxygenase | |
| 50 | Bpet1331 | 1393394..1395133 [+], 1740 | 2,4-dichlorophenol 6-monooxygenase | |
| 51 | Bpet1332 | 1395199..1395825 [+], 627 | transcriptional regulator, TetR-family | |
| 52 | Bpet1333 | 1395907..1397118 [+], 1212 | permease of the major facilitator family | |
| 53 | Bpet1334 | 1397137..1397862 [+], 726 | hypothetical Protein | |
| 54 | Bpet1335 | 1397873..1398595 [-], 723 | conserved hypothetical protein | |
| 55 | Bpet1336 | 1398784..1399206 [-], 423 | conserved hypothetical protein, fragment | |
| 56 | tnp6 | 1399285..1399572 [+], 288 | transposase | |
| 57 | Bpet1338 | 1399569..1400459 [+], 891 | probable transposase | |
| 58 | Bpet1339 | 1400498..1400833 [-], 336 | hypothetical protein predicted by Glimmer/Critica | |
| 59 | Bpet1340 | 1400898..1401554 [-], 657 | Threonine efflux protein. | |
| 60 | Bpet1341 | 1401556..1402326 [-], 771 | conserved hypothetical protein | |
| 61 | alaS1 | 1402426..1403058 [-], 633 | alanyl-tRNA synthetase domain protein | |
| 62 | Bpet1343 | 1403154..1404140 [+], 987 | transcriptional regulator, LysR-family | |
| 63 | Bpet1344 | 1404231..1404815 [+], 585 | putative lipoprotein | Tfc2, T4SS component |
| 64 | Bpet1345 | 1404812..1405459 [+], 648 | putative membrane protein | Tfc2, T4SS component |
| 65 | Bpet1346 | 1405469..1406209 [+], 741 | conserved protein, putatively exported | Tfc3, T4SS component |
| 66 | Bpet1347 | 1406194..1406778 [+], 585 | conserved exported protein | Tfc4, T4SS component |
| 67 | Bpet1348 | 1406775..1407326 [+], 552 | putative secreted protein | Tfc5, T4SS component |
| 68 | Bpet1349 | 1407337..1409505 [+], 2169 | conserved hypothetical protein | Tfc6, T4SS component |
| 69 | Bpet1350 | 1409502..1410251 [+], 750 | putative membrane protein | Tfc8, T4SS component |
| 70 | csrA | 1410532..1410729 [+], 198 | carbon storage regulator | |
| 71 | Bpet1352 | 1410780..1412069 [-], 1290 | conserved hypothetical protein | |
| 72 | Bpet1353 | 1412066..1412434 [-], 369 | putative DNA-binding protein | |
| 73 | Bpet1354 | 1412789..1413145 [+], 357 | putative secreted protein | Tfc9, T4SS component |
| 74 | Bpet1355 | 1413142..1413375 [+], 234 | - | |
| 75 | Bpet1356 | 1413392..1413757 [+], 366 | conserved hypothetical protein | Tfc10, T4SS component |
| 76 | Bpet1357 | 1413767..1414168 [+], 402 | putative membrane protein | Tfc11, T4SS component |
| 77 | Bpet1358 | 1414165..1414857 [+], 693 | conserved hypothetical protein | Tfc12, T4SS component |
| 78 | Bpet1359 | 1414854..1415762 [+], 909 | conserved hypothetical protein | Tfc13, T4SS component |
| 79 | Bpet1360 | 1415752..1417200 [+], 1449 | putative exported protein | Tfc14, T4SS component |
| 80 | Bpet1361 | 1417181..1417615 [+], 435 | putative lipoprotein | Tfc15, T4SS component |
| 81 | Bpet1362 | 1417615..1420494 [+], 2880 | conserved hypothetical protein | Tfc16, T4SS component |
| 82 | Bpet1363 | 1420550..1422661 [-], 2112 | probable signalling protein | |
| 83 | Bpet1364 | 1423188..1423943 [+], 756 | conserved hypothetical protein | |
| 84 | Bpet1365 | 1423971..1424603 [-], 633 | conserved hypothetical protein | |
| 85 | Bpet1366 | 1425030..1426340 [-], 1311 | conserved hypothetical protein | |
| 86 | Bpet1367 | 1426282..1427076 [-], 795 | transcriptional regulator, TetR-family | |
| 87 | Bpet1368 | 1427315..1429882 [-], 2568 | probable signalling protein | |
| 88 | Bpet1369 | 1430684..1432189 [+], 1506 | - | |
| 89 | Bpet1370 | 1432239..1433117 [-], 879 | transcriptional regulator, LysR-family | |
| 90 | Bpet1371 | 1433241..1434017 [+], 777 | hypothetical protein | |
| 91 | rhtB2 | 1434074..1434691 [+], 618 | putative amino acid efflux transmembrane protein | |
| 92 | Bpet1373 | 1434704..1435333 [+], 630 | - | |
| 93 | Bpet1374 | 1435548..1435997 [+], 450 | conserved hypothetical protein | Tfc24, T4SS component |
| 94 | Bpet1375 | 1435994..1436938 [+], 945 | conserved hypothetical protein | Tfc23, T4SS component |
| 95 | Bpet1376 | 1436940..1438346 [+], 1407 | conserved hypothetical protein | Tfc22, T4SS component |
| 96 | Bpet1377 | 1438343..1438693 [+], 351 | conserved hypothetical protein | |
| 97 | Bpet1378 | 1438707..1440224 [+], 1518 | conserved hypothetical protein | Tfc19, T4SS component |
| 98 | Bpet1379 | 1440233..1440610 [-], 378 | hypothetical protein | |
| 99 | Bpet1380 | 1440673..1441341 [-], 669 | conserved hypothetical protein | |
| 100 | Bpet1381 | 1441406..1441654 [-], 249 | hypothetical protein predicted by Glimmer/Critica | |
| 101 | Bpet1382 | 1442143..1443972 [+], 1830 | putative helicase | Relaxase, MOBH Family |
| 102 | Bpet1383 | 1444018..1445601 [-], 1584 | conserved hypothetical protein | |
| 103 | Bpet1384 | 1445598..1446872 [-], 1275 | conserved hypothetical protein | |
| 104 | benE | 1446869..1448050 [-], 1182 | benzoate membrane transport protein | |
| 105 | Bpet1386 | 1448078..1449436 [-], 1359 | putative n-hydroxybenzoate transporter | |
| 106 | areB | 1449517..1450647 [-], 1131 | Aryl-alcohol dehydrogenase | |
| 107 | Bpet1388 | 1450658..1452124 [-], 1467 | benzaldehyde dehydrogenase II | |
| 108 | Bpet1389 | 1452184..1452570 [-], 387 | Hypothetical UPF0065 protein in clcB-clcD intergenic region precursor. | |
| 109 | Bpet1390 | 1452695..1452973 [-], 279 | methylmuconolactone isomerase | |
| 110 | Bpet1391 | 1453007..1453819 [-], 813 | 3-oxoadipate enol-lactone hydrolase | |
| 111 | Bpet1392 | 1453830..1454108 [-], 279 | putative thiolase, fragment | |
| 112 | Bpet1393 | 1454092..1454286 [-], 195 | 3-oxoadipate CoA-transferase subunit A, fragment | |
| 113 | benK | 1454310..1455671 [-], 1362 | benzoate transport protein, putative | |
| 114 | Bpet1395 | 1455709..1456125 [-], 417 | conserved hypothetical protein | |
| 115 | benD | 1456182..1456955 [-], 774 | cis-1,2-dihydroxycyclohexa-3, 5-diene-1-carboxylate dehydrogenase,(cis-1,2-dihydroxy-3, 4-cyclohexadiene-1-carboxylate dehydrogenase) | |
| 116 | benC | 1456968..1457993 [-], 1026 | Ferredoxin--NAD(+) reductase | |
| 117 | benB | 1458011..1458511 [-], 501 | benzoate 1,2-dioxygenase | |
| 118 | benA | 1458508..1459872 [-], 1365 | Benzoate 1,2-dioxygenase alpha subunit | |
| 119 | catA2 | 1459999..1460934 [-], 936 | catechol 1,2-dioxygenase | |
| 120 | catB1 | 1460977..1462197 [-], 1221 | chloromuconate cycloisomerase | |
| 121 | catR1 | 1462273..1463271 [+], 999 | transcriptional regulator catR | |
| 122 | Bpet1403 | 1463665..1464102 [+], 438 | putative transposase | |
| 123 | Bpet1404 | 1464072..1464458 [+], 387 | transposase IS4 family | |
| 124 | Bpet1405 | 1464541..1464735 [-], 195 | putative transposase A | |
| 125 | Bpet1406 | 1464809..1465105 [-], 297 | putative transposase | |
| 126 | Bpet1407 | 1465132..1465323 [+], 192 | conserved hypothetical protein | |
| 127 | Bpet1408 | 1465555..1466010 [+], 456 | conserved hypothetical protein | |
| 128 | Bpet1409 | 1466321..1468411 [+], 2091 | transposase | |
| 129 | Bpet1410 | 1468264..1469049 [-], 786 | integrase, catalytic domain | |
| 130 | Bpet1411 | 1469103..1469429 [-], 327 | transposase IS911 HTH and LZ region | |
| 131 | Bpet1412 | 1469516..1470343 [-], 828 | hypothetical protein | |
| 132 | Bpet1413 | 1470523..1470714 [-], 192 | conserved hypothetical protein | |
| 133 | Bpet1414 | 1470755..1471189 [-], 435 | conserved hypothetical protein | |
| 134 | nthB | 1471192..1471848 [-], 657 | nitrile hydratase subunit beta (Nitrilase) (NHase) | |
| 135 | nthA | 1471859..1472620 [-], 762 | nitrile hydratase alpha subunit | |
| 136 | Bpet1417 | 1472754..1474160 [-], 1407 | glutamyl-tRNA(Gln) amidotransferase subunit A | |
| 137 | Bpet1418 | 1474157..1474363 [-], 207 | hypothetical protein predicted by Glimmer/Critica | |
| 138 | Bpet1419 | 1474894..1475280 [+], 387 | transposase IS4 family | |
| 139 | Bpet1420 | 1475915..1477492 [-], 1578 | - | |
| 140 | Bpet1421 | 1478386..1478928 [+], 543 | hypothetical protein predicted by Glimmer/Critica | |
| 141 | Bpet1422 | 1479015..1479320 [+], 306 | hypothetical protein predicted by Glimmer/Critica | |
| 142 | Bpet1423 | 1479382..1479675 [-], 294 | hypothetical protein predicted by Glimmer/Critica | |
| 143 | Bpet1424 | 1480368..1480523 [+], 156 | IS5 family element, transposase orfA | |
| 144 | Bpet1425 | 1480737..1481072 [+], 336 | 3-oxoacid CoA-transferase subunit A, fragment | |
| 145 | Bpet1426 | 1481151..1481471 [+], 321 | 3-oxoacid CoA-transferase subunit B, fragment | |
| 146 | Bpet1427 | 1481766..1482545 [+], 780 | transcriptional regulatory protein | |
| 147 | Bpet1428 | 1482530..1483702 [-], 1173 | putative salicylate hydroxylase | |
| 148 | Bpet1429 | 1483719..1484573 [-], 855 | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase | |
| 149 | Bpet1430 | 1484603..1485646 [-], 1044 | putative gentisate 1,2-dioxygenase | |
| 150 | Bpet1431 | 1485814..1487190 [+], 1377 | putative 4-hydroxybenzoate transporter | |
| 151 | tnp7 | 1487225..1487650 [-], 426 | putative resolvase | |
| 152 | Bpet1433 | 1488052..1488378 [-], 327 | putative transcriptional regulator | |
| 153 | Bpet1434 | 1488485..1489135 [+], 651 | probable transcriptional regulator | |
| 154 | Bpet1435 | 1489427..1490008 [+], 582 | transcriptional regulator, TetR-family | |
| 155 | Bpet1436 | 1490093..1491262 [+], 1170 | putative Transmembrane efflux protein of the MFS type | |
| 156 | intB13 | 1491479..1493314 [-], 1836 | phage-related integrase | |
| 157 | Bpet1438 | 1494345..1495100 [+], 756 | conserved hypothetical protein | |
| 158 | alpA3 | 1495220..1495432 [+], 213 | predicted transcriptional regulator (phage-related) | |
| 159 | parA3 | 1495476..1496351 [+], 876 | hypothetical protein | |
| 160 | Bpet1441 | 1496335..1496592 [+], 258 | conserved hypothetical protein | |
| 161 | Bpet1442 | 1496585..1498237 [+], 1653 | conserved hypothetical protein | |