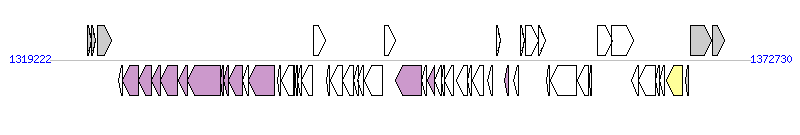The graph information of ICE(Tn4371)6068 components from AM743169 |
 |
| Complete gene list of ICE(Tn4371)6068 from AM743169 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | rplU | 1322082..1322390 [+], 309 | putative 50S ribosomal protein L21 | |
| 2 | rpmA | 1322409..1322672 [+], 264 | putative 50S ribosomal protein L27 | |
| 3 | Smlt1280 | 1322866..1323918 [+], 1053 | putative conserved GTP-binding protein | |
| 4 | Smlt1282 | 1324492..1324737 [-], 246 | conserved hypothetical protein | |
| 5 | trbI | 1324734..1325993 [-], 1260 | putative conjugal transfer protein | TrbI, T4SS component |
| 6 | trbG | 1325996..1326982 [-], 987 | putative conjugal transfer protein | TrbG, T4SS component |
| 7 | trbF | 1326979..1327683 [-], 705 | putative conjugal transfer protein | TrbF, T4SS component |
| 8 | trbL | 1327702..1329000 [-], 1299 | putative conjugal transfer protein | TrbL, T4SS component |
| 9 | trbJ | 1329012..1329749 [-], 738 | putative conjugal transfer protein | TrbJ, T4SS component |
| 10 | trbE | 1329746..1332232 [-], 2487 | putative conjugal transfer protein | TrbE, T4SS component |
| 11 | trbD | 1332243..1332515 [-], 273 | putative conjugal transfer protein | TrbD, T4SS component |
| 12 | trbC | 1332512..1332895 [-], 384 | putative conjugal transfer protein | TrbC, T4SS component |
| 13 | trbB | 1332892..1333932 [-], 1041 | putative conjugal transfer protein | TrbB, T4SS component |
| 14 | Smlt1292 | 1333929..1334375 [-], 447 | conserved hypothetical protein | |
| 15 | traG | 1334372..1336372 [-], 2001 | putative conjugal transfer protein | VirD4, T4SS component |
| 16 | Smlt1294 | 1336606..1336866 [-], 261 | conserved hypothetical protein | |
| 17 | Smlt1295 | 1336911..1337804 [-], 894 | putative LysR family transcriptional regulator | |
| 18 | Smlt1296 | 1337815..1337940 [-], 126 | putative regulator, pseudogene | |
| 19 | Smlt1297 | 1337956..1338126 [-], 171 | putative esterase, pseudogene | |
| 20 | Smlt1298 | 1338123..1338425 [-], 303 | conserved hypothetical protein | |
| 21 | Smlt1299 | 1338412..1339266 [-], 855 | putative NAD(P)H dehydrogenase [quinone] | |
| 22 | Smlt1300 | 1339362..1340264 [+], 903 | putative LysR family transcriptional regulator | |
| 23 | Smlt1301 | 1340359..1340619 [-], 261 | conserved hypothetical protein | |
| 24 | Smlt1302 | 1340656..1341549 [-], 894 | putative LysR family transcriptional regulator | |
| 25 | Smlt1303 | 1341582..1342424 [-], 843 | conserved hypothetical protein | |
| 26 | Smlt1304 | 1342421..1342804 [-], 384 | putative 4-carboxymuconolactone decarboxylase | |
| 27 | Smlt1305 | 1342737..1343207 [-], 471 | putative MerR family transcriptional regulator | |
| 28 | Smlt1306 | 1343295..1344665 [-], 1371 | putative MFS family transmembrane transporter | |
| 29 | Smlt1307 | 1344784..1345635 [+], 852 | putative LysR family transcriptional regulator | |
| 30 | Smlt1308 | 1345632..1347596 [-], 1965 | conserved hypothetical protein | Relaxase, MOBL Family |
| 31 | Smlt1309 | 1347593..1348030 [-], 438 | conserved hypothetical protein | |
| 32 | Smlt1310 | 1348056..1348631 [-], 576 | putative transmembrane anchor conjugal transfer protein | TraF, T4SS component |
| 33 | Smlt1311 | 1348628..1349152 [-], 525 | conserved hypothetical protein | |
| 34 | Smlt1312 | 1349149..1349412 [-], 264 | putative parB partition protein | |
| 35 | Smlt1313 | 1349409..1350047 [-], 639 | putative ParA/CobQ/CobB/MinD nucleotide binding domain protein | |
| 36 | Smlt1314 | 1350298..1351131 [-], 834 | putative RepA-like replication protein | |
| 37 | Smlt1315 | 1351158..1351448 [-], 291 | conserved hypothetical protein | |
| 38 | Smlt1316 | 1351532..1352335 [-], 804 | conserved hypothetical protein | |
| 39 | Smlt1318 | 1352687..1353037 [-], 351 | conserved hypothetical protein | |
| 40 | Smlt1319 | 1353340..1353636 [+], 297 | putative HTH transcriptional regulator | |
| 41 | Smlt1320 | 1353993..1354307 [-], 315 | conserved hypothetical protein | TraF, T4SS component |
| 42 | Smlt1320A | 1354642..1355064 [-], 423 | conserved hypothetical protein | |
| 43 | Smlt1322 | 1355155..1355490 [+], 336 | hypothetical protein | |
| 44 | Smlt1323 | 1355532..1356575 [+], 1044 | putative transposase | |
| 45 | Smlt1324 | 1356572..1357087 [+], 516 | conserved hypothetical protein | |
| 46 | Smlt1325 | 1357200..1357388 [-], 189 | conserved hypothetical protein | |
| 47 | Smlt1326 | 1357451..1359481 [-], 2031 | putative ParB-like nuclease domain protein | |
| 48 | Smlt1327 | 1359562..1360392 [-], 831 | conserved hypothetical protein | |
| 49 | Smlt1328 | 1360425..1360622 [-], 198 | conserved hypothetical protein | |
| 50 | Smlt1329 | 1361053..1362183 [+], 1131 | conserved hypothetical protein | |
| 51 | Smlt1331 | 1362149..1363831 [+], 1683 | putative transmembrane protein | |
| 52 | Smlt1332 | 1363664..1364188 [-], 525 | putative RadC DNA repair protein, pseudogene | |
| 53 | Smlt1333 | 1364225..1365508 [-], 1284 | conserved hypothetical protein | |
| 54 | Smlt1334 | 1365534..1365839 [-], 306 | conserved hypothetical protein | |
| 55 | Smlt1335 | 1365820..1366203 [-], 384 | conserved hypothetical protein | |
| 56 | Smlt1336 | 1366343..1367584 [-], 1242 | putative prophage integrase | Integrase |
| 57 | rpsT | 1367795..1368064 [-], 270 | putative 30S ribosomal protein S20 | |
| 58 | mviN | 1368167..1369786 [+], 1620 | putative transmembrane virulence factor MmviN | |
| 59 | ribF | 1369862..1370809 [+], 948 | putative riboflavin biosynthesis protein RibF | |