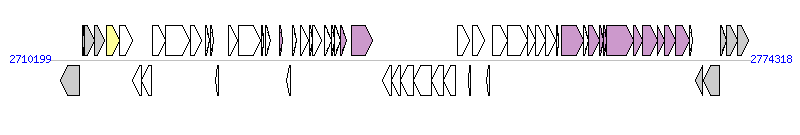The graph information of ICE(Tn4371)6033 components from CP001068 |
 |
| Complete gene list of ICE(Tn4371)6033 from CP001068 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | Rpic_2597 | 2711130..2712917 [-], 1788 | surface antigen (D15) | |
| 2 | Rpic_2598 | 2713155..2713343 [+], 189 | conserved hypothetical protein | |
| 3 | Rpic_2599 | 2713383..2714264 [+], 882 | chromosome segregation and condensation protein ScpA | |
| 4 | Rpic_2600 | 2714303..2715154 [+], 852 | pantoate/beta-alanine ligase | |
| 5 | Rpic_2601 | 2715382..2716581 [+], 1200 | integrase family protein | Integrase |
| 6 | Rpic_2602 | 2716578..2717714 [+], 1137 | protein of unknown function DUF1016 | |
| 7 | Rpic_2603 | 2717760..2718548 [-], 789 | conserved hypothetical protein | |
| 8 | Rpic_2604 | 2718555..2719457 [-], 903 | conserved hypothetical protein | |
| 9 | Rpic_2605 | 2719547..2720740 [+], 1194 | conserved hypothetical protein | |
| 10 | Rpic_2606 | 2720737..2722992 [+], 2256 | UBA/THIF-type NAD/FAD binding protein | |
| 11 | Rpic_2607 | 2723038..2724108 [+], 1071 | conserved hypothetical protein | |
| 12 | Rpic_2608 | 2724485..2724835 [+], 351 | conserved hypothetical protein | |
| 13 | Rpic_2609 | 2724899..2725177 [+], 279 | conserved hypothetical protein | |
| 14 | Rpic_2610 | 2725320..2725619 [-], 300 | conserved hypothetical protein | |
| 15 | Rpic_2611 | 2726555..2727379 [+], 825 | protein of unknown function DUF932 | |
| 16 | Rpic_2612 | 2727445..2729484 [+], 2040 | ParB domain protein nuclease | |
| 17 | Rpic_2613 | 2729547..2729756 [+], 210 | conserved hypothetical protein | |
| 18 | Rpic_2614 | 2729973..2730434 [+], 462 | conserved hypothetical protein | |
| 19 | Rpic_2615 | 2731222..2731536 [+], 315 | protein of unknown function DUF736 | TraF, T4SS component |
| 20 | Rpic_2616 | 2731902..2732195 [-], 294 | helix-turn-helix domain protein | |
| 21 | Rpic_2617 | 2732419..2732766 [+], 348 | putative lipoprotein | |
| 22 | Rpic_2618 | 2733108..2733857 [+], 750 | conserved hypothetical protein | |
| 23 | Rpic_2619 | 2733941..2734222 [+], 282 | conserved hypothetical protein | |
| 24 | Rpic_2620 | 2734249..2735097 [+], 849 | conserved hypothetical replication initiator and transcription repressor protein | |
| 25 | Rpic_2621 | 2735351..2735989 [+], 639 | Cobyrinic acid ac-diamide synthase | |
| 26 | Rpic_2622 | 2735986..2736249 [+], 264 | conserved hypothetical protein | |
| 27 | Rpic_2623 | 2736246..2736779 [+], 534 | conserved hypothetical protein | |
| 28 | Rpic_2624 | 2736776..2737363 [+], 588 | putative conjugal transfer TRAF transmembrane protein | TraF, T4SS component |
| 29 | Rpic_2625 | 2737800..2739782 [+], 1983 | conserved hypothetical protein | Relaxase, MOBL Family |
| 30 | Rpic_2627 | 2740659..2741441 [-], 783 | short-chain dehydrogenase/reductase SDR | |
| 31 | Rpic_2628 | 2741476..2742261 [-], 786 | Enoyl-CoA hydratase/isomerase | |
| 32 | Rpic_2629 | 2742273..2743454 [-], 1182 | acetyl-CoA acetyltransferase | |
| 33 | Rpic_2630 | 2743483..2745159 [-], 1677 | AMP-dependent synthetase and ligase | |
| 34 | Rpic_2631 | 2745171..2746220 [-], 1050 | L-carnitine dehydratase/bile acid-inducible protein F | |
| 35 | Rpic_2632 | 2746223..2747368 [-], 1146 | acyl-CoA dehydrogenase domain protein | |
| 36 | Rpic_2633 | 2747563..2748591 [+], 1029 | transcriptional regulator, AraC family | |
| 37 | Rpic_2634 | 2748564..2748671 [-], 108 | hypothetical protein | |
| 38 | Rpic_2635 | 2748939..2749988 [+], 1050 | beta-lactamase domain protein | |
| 39 | Rpic_2636 | 2750177..2750455 [-], 279 | transposase family protein | |
| 40 | Rpic_2637 | 2750723..2752090 [+], 1368 | major facilitator superfamily MFS_1 | |
| 41 | Rpic_2638 | 2752115..2753896 [+], 1782 | acyl-CoA dehydrogenase domain protein | |
| 42 | Rpic_2639 | 2753921..2754670 [+], 750 | Electron transfer flavoprotein alpha/beta-subunit | |
| 43 | Rpic_2640 | 2754671..2755603 [+], 933 | Electron transfer flavoprotein alpha subunit | |
| 44 | Rpic_2641 | 2755621..2756535 [+], 915 | transcriptional regulator, LysR family | |
| 45 | Rpic_2642 | 2756561..2756791 [+], 231 | conserved hypothetical protein | |
| 46 | Rpic_2643 | 2757009..2759027 [+], 2019 | TRAG family protein | VirD4, T4SS component |
| 47 | Rpic_2644 | 2759024..2759488 [+], 465 | CopG domain protein DNA-binding domain protein | |
| 48 | Rpic_2645 | 2759485..2760522 [+], 1038 | P-type conjugative transfer ATPase TrbB | TrbB, T4SS component |
| 49 | Rpic_2646 | 2760519..2760911 [+], 393 | Conjugal transfer protein TrbC | TrbC, T4SS component |
| 50 | Rpic_2647 | 2760908..2761192 [+], 285 | putative conjugal transfer TrbD transmembrane protein | TrbD, T4SS component |
| 51 | Rpic_2648 | 2761212..2763689 [+], 2478 | AAA ATPase | TrbE, T4SS component |
| 52 | Rpic_2649 | 2763686..2764441 [+], 756 | P-type conjugative transfer protein TrbJ | TrbJ, T4SS component |
| 53 | Rpic_2650 | 2764454..2765806 [+], 1353 | P-type conjugative transfer protein TrbL | TrbL, T4SS component |
| 54 | Rpic_2651 | 2765825..2766529 [+], 705 | Conjugal transfer protein | TrbF, T4SS component |
| 55 | Rpic_2652 | 2766526..2767521 [+], 996 | P-type conjugative transfer protein TrbG | TrbG, T4SS component |
| 56 | Rpic_2653 | 2767524..2768795 [+], 1272 | conjugation TrbI family protein | TrbI, T4SS component |
| 57 | Rpic_2654 | 2768792..2769037 [+], 246 | conserved hypothetical protein | |
| 58 | Rpic_2655 | 2769315..2769980 [-], 666 | putative partition-related protein | |
| 59 | Rpic_2656 | 2770055..2771551 [-], 1497 | cobyric acid synthase CobQ | |
| 60 | Rpic_2657 | 2771613..2772188 [+], 576 | Adenosylcobinamide-phosphate guanylyltransferase | |
| 61 | Rpic_2658 | 2772185..2773165 [+], 981 | cobalamin biosynthesis protein CobD | |
| 62 | Rpic_2659 | 2773162..2774169 [+], 1008 | L-threonine-O-3-phosphate decarboxylase | |