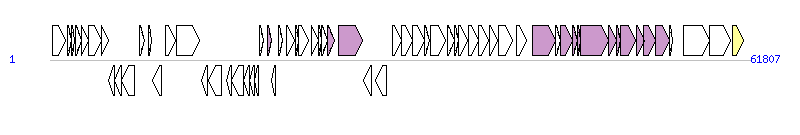The graph information of ICE-KKS components from AB546270 |
 |
| Complete gene list of ICE-KKS from AB546270 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 186..1388 [+], 1203 | integrase | |
| 2 | - | 1528..1911 [+], 384 | hypothetical protein | |
| 3 | - | 1895..2197 [+], 303 | hypothetical protein | |
| 4 | - | 2286..2753 [+], 468 | hypothetical protein | |
| 5 | - | 2753..3379 [+], 627 | hypothetical protein | |
| 6 | - | 3383..4519 [+], 1137 | hypothetical protein | |
| 7 | - | 4516..5178 [+], 663 | hypothetical protein | |
| 8 | - | 5195..5680 [-], 486 | hypothetical protein | |
| 9 | - | 5722..6309 [-], 588 | hypothetical protein | |
| 10 | - | 6313..7455 [-], 1143 | hypothetical protein | |
| 11 | radC | 7868..8374 [+], 507 | DNA repair protein RadC | |
| 12 | - | 8656..9006 [+], 351 | hypothetical protein | |
| 13 | - | 9037..9816 [-], 780 | hypothetical protein | |
| 14 | gene32 | 10233..11057 [+], 825 | putative F plasmid gene 32-like protein | |
| 15 | parB | 11136..13196 [+], 2061 | ParB family protein | |
| 16 | - | 13404..13934 [-], 531 | hypothetical protein | |
| 17 | - | 14017..15168 [-], 1152 | chromate transporter | |
| 18 | arsC | 15566..15988 [-], 423 | arsenate reductase | |
| 19 | - | 16006..17091 [-], 1086 | arsenite transporter | |
| 20 | - | 17102..17599 [-], 498 | hypothetical protein | |
| 21 | - | 17612..18082 [-], 471 | Glyoxalase/bleomycin resistance protein | |
| 22 | - | 18096..18443 [-], 348 | ArsR family transcriptional regulator | |
| 23 | - | 18516..18875 [+], 360 | hypothetical protein | |
| 24 | - | 19172..19525 [+], 354 | hypothetical protein | TraF, T4SS component |
| 25 | - | 19591..19896 [-], 306 | XRE family transcriptional regulator | |
| 26 | - | 20137..20487 [+], 351 | Putative lipoprotein | |
| 27 | - | 20867..21571 [+], 705 | hypothetical protein | |
| 28 | xis | 21656..21937 [+], 282 | excisionase | |
| 29 | rep | 21964..22821 [+], 858 | plasmid replication protein | |
| 30 | parA | 23075..23713 [+], 639 | plasmid partitioning protein | |
| 31 | - | 23710..23994 [+], 285 | hypothetical protein | |
| 32 | - | 23991..24524 [+], 534 | hypothetical protein | |
| 33 | traF | 24521..25117 [+], 597 | conjugal transfer protein TraF | TraF, T4SS component |
| 34 | traI | 25452..27551 [+], 2100 | relaxase | Relaxase, MOBL Family |
| 35 | bphS | 27659..28423 [-], 765 | GntR family transcriptional regulator | |
| 36 | tnp | 28778..29743 [-], 966 | transposase | |
| 37 | bphE | 30255..31061 [+], 807 | 2-hydroxypent-2,4-dienoate hydratase | |
| 38 | bphG | 31064..32014 [+], 951 | acetaldehyde dehydrogenase | |
| 39 | bphF | 32025..33083 [+], 1059 | 4-hydroxy-2-oxovalerate aldolase | |
| 40 | bphV | 33102..33512 [+], 411 | putative membrane protein | |
| 41 | bphA1 | 33572..34948 [+], 1377 | biphenyl dioxygenase alpha subunit | |
| 42 | bphA2 | 35119..35700 [+], 582 | biphenyl dioxygenase beta subunit | |
| 43 | bphA3 | 35718..36047 [+], 330 | biphenyl dioxygenase ferredoxin subunit | |
| 44 | bphB | 36066..36896 [+], 831 | cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase | |
| 45 | bphC | 36911..37792 [+], 882 | dihydroxybiphenyl dioxygenase | |
| 46 | bphD | 37829..38689 [+], 861 | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid hydrolase | |
| 47 | bphW | 38817..39473 [+], 657 | putative outer membrane protein | |
| 48 | bphA4 | 39638..40864 [+], 1227 | biphenyl dioxygenase ferredoxin reductase subunit | |
| 49 | traR | 41158..42102 [+], 945 | LysR family transcriptional regulator | |
| 50 | traG | 42613..44640 [+], 2028 | conjugal transfer coupling protein TraG | VirD4, T4SS component |
| 51 | - | 44637..45113 [+], 477 | hypothetical protein | |
| 52 | trbB | 45110..46177 [+], 1068 | conjugal transfer protein TrbB | TrbB, T4SS component |
| 53 | trbC | 46174..46557 [+], 384 | conjugal transfer protein TrbC | TrbC, T4SS component |
| 54 | trbD | 46554..46826 [+], 273 | conjugal transfer protein TrbD | TrbD, T4SS component |
| 55 | trbE | 46842..49295 [+], 2454 | conjugal transfer protein TrbE | TrbE, T4SS component |
| 56 | trbJ | 49292..50017 [+], 726 | conjugal transfer protein TrbJ | TrbJ, T4SS component |
| 57 | - | 50030..50350 [+], 321 | lipoprotein | TrbJ, T4SS component |
| 58 | trbL | 50347..51732 [+], 1386 | conjugal transfer protein TrbL | TrbL, T4SS component |
| 59 | trbF | 51745..52449 [+], 705 | conjugal transfer protein TrbF | TrbF, T4SS component |
| 60 | trbG | 52446..53429 [+], 984 | conjugal transfer protein TrbG | TrbG, T4SS component |
| 61 | trbI | 53432..54718 [+], 1287 | conjugal transfer protein TrbI | TrbI, T4SS component |
| 62 | - | 54715..54933 [+], 219 | hypothetical protein | |
| 63 | - | 55918..58254 [+], 2337 | hypothetical protein | |
| 64 | - | 58251..60128 [+], 1878 | DNA helicase | |
| 65 | tnp | 60283..61230 [+], 948 | transposase | Integrase |