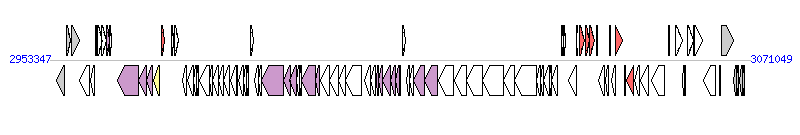The graph information of ICEVchBan9 components from CP001485 |
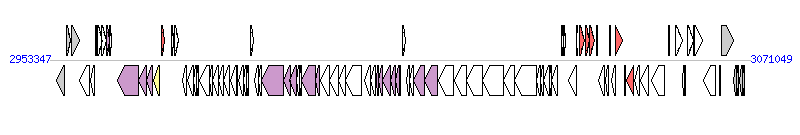 |
| Complete gene list of ICEVchBan9 from CP001485 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | VCD_003657 | 2954522..2955799 [-], 1278 | branched-chain amino acid transport system carrier protein | |
| 2 | VCD_003658 | 2956144..2956866 [+], 723 | predicted O-methyltransferase | |
| 3 | VCD_003659 | 2956954..2958225 [+], 1272 | ATP-dependent RNA helicase SrmB | |
| 4 | VCD_003660 | 2958347..2959936 [-], 1590 | peptide chain release factor 3 | |
| 5 | VCD_003661 | 2960193..2960840 [-], 648 | HTH-type transcriptional regulator for conjugative element SXT | |
| 6 | VCD_003662 | 2960958..2961209 [+], 252 | uncharacterized protein conserved in bacteria prophage-related | |
| 7 | VCD_003663 | 2961265..2962134 [+], 870 | hypothetical protein | |
| 8 | VCD_003664 | 2962055..2962783 [+], 729 | hypothetical protein | |
| 9 | VCD_003665 | 2962770..2963318 [+], 549 | soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) | Orf169_F, T4SS component |
| 10 | VCD_003666 | 2963315..2963614 [+], 300 | hypothetical protein | |
| 11 | VCD_003667 | 2964666..2968235 [-], 3570 | IncF plasmid conjugative transfer protein TraG | TraG_F, T4SS component |
| 12 | VCD_003668 | 2968239..2969627 [-], 1389 | IncF plasmid conjugative transfer pilus assembly protein TraH | TraH_F, T4SS component |
| 13 | VCD_003669 | 2969630..2970574 [-], 945 | IncF plasmid conjugative transfer pilus assembly protein TraF | TraF_F, T4SS component |
| 14 | VCD_003670 | 2970831..2971790 [-], 960 | integron integrase | Integrase |
| 15 | VCD_003671 | 2972078..2972551 [+], 474 | dihydrofolate reductase | AR |
| 16 | VCD_003672 | 2973754..2974122 [+], 369 | hypothetical protein | |
| 17 | VCD_003673 | 2974222..2974923 [+], 702 | hypothetical protein | |
| 18 | VCD_003674 | 2975596..2976303 [-], 708 | hypothetical protein | |
| 19 | VCD_003675 | 2976393..2977466 [-], 1074 | hypothetical protein | |
| 20 | VCD_003676 | 2977557..2977898 [-], 342 | hypothetical protein | |
| 21 | VCD_003677 | 2977898..2978395 [-], 498 | DNA repair protein RadC | |
| 22 | VCD_003678 | 2978480..2980135 [-], 1656 | hypothetical protein | |
| 23 | VCD_003679 | 2980205..2980645 [-], 441 | hypothetical protein | |
| 24 | VCD_003680 | 2980707..2981660 [-], 954 | cobalamine biosynthesis protein | |
| 25 | VCD_003681 | 2981759..2982529 [-], 771 | hypothetical protein | |
| 26 | VCD_003682 | 2982526..2983602 [-], 1077 | ATPase associated with various cellular activities AAA_5 | |
| 27 | VCD_003683 | 2983695..2984711 [-], 1017 | DNA recombination protein | |
| 28 | VCD_003684 | 2984772..2984915 [-], 144 | hypothetical protein | |
| 29 | VCD_003685 | 2984997..2985815 [-], 819 | hypothetical protein | |
| 30 | VCD_003686 | 2985895..2986314 [-], 420 | single-stranded DNA-binding protein | |
| 31 | VCD_003687 | 2986330..2986668 [-], 339 | hypothetical protein | |
| 32 | VCD_003688 | 2987024..2987626 [+], 603 | hypothetical protein | |
| 33 | VCD_003689 | 2987717..2988379 [-], 663 | hypothetical protein | |
| 34 | VCD_003690 | 2988410..2988883 [-], 474 | hypothetical protein | |
| 35 | VCD_003691 | 2988974..2992615 [-], 3642 | IncF plasmid conjugative transfer protein TraN | TraN_F, T4SS component |
| 36 | VCD_003692 | 2992669..2993703 [-], 1035 | IncF plasmid conjugative transfer pilus assembly protein TraU | TraU_F, T4SS component |
| 37 | VCD_003693 | 2993681..2994844 [-], 1164 | hypothetical protein | TraW_F, T4SS component |
| 38 | VCD_003694 | 2994816..2995328 [-], 513 | signal peptidase I | |
| 39 | VCD_003695 | 2995312..2995659 [-], 348 | hypothetical protein | |
| 40 | VCD_003696 | 2995652..2998069 [-], 2418 | IncF plasmid conjugative transfer pilus assembly protein TraC | TraC_F, T4SS component |
| 41 | VCD_003697 | 2998051..2998743 [-], 693 | thiol:disulfide interchange protein DsbC | TrbB_I, T4SS component |
| 42 | VCD_003698 | 2998888..3000333 [-], 1446 | type 4 fimbriae expression regulatory protein pilR | |
| 43 | VCD_003699 | 3000336..3001871 [-], 1536 | hypothetical protein | |
| 44 | VCD_003700 | 3001881..3002990 [-], 1110 | hypothetical protein | |
| 45 | VCD_003701 | 3002987..3005716 [-], 2730 | hypothetical protein | |
| 46 | VCD_003702 | 3006281..3007231 [-], 951 | Ync | |
| 47 | VCD_003703 | 3007212..3008045 [-], 834 | Ynd | |
| 48 | VCD_003704 | 3008223..3008609 [-], 387 | TraA | TraA_F, T4SS component |
| 49 | VCD_003705 | 3008606..3009256 [-], 651 | conjugative transfer protein TraV | TraV_F, T4SS component |
| 50 | VCD_003706 | 3009253..3010545 [-], 1293 | IncF plasmid conjugative transfer pilus assembly protein TraB | TraB_F, T4SS component |
| 51 | VCD_003707 | 3010542..3011444 [-], 903 | IncF plasmid conjugative transfer pilus assembly protein TraK | TraK_F, T4SS component |
| 52 | VCD_003708 | 3011425..3012051 [-], 627 | hypothetical protein | TraE_F, T4SS component |
| 53 | VCD_003709 | 3012048..3012335 [-], 288 | IncF plasmid conjugative transfer pilus assembly protein TraL | TraL_F, T4SS component |
| 54 | VCD_003710 | 3012585..3013205 [+], 621 | hypothetical protein | |
| 55 | VCD_003711 | 3013232..3013876 [-], 645 | conjugative transfer protein s043 | |
| 56 | VCD_003712 | 3013854..3014390 [-], 537 | hypothetical protein | |
| 57 | VCD_003713 | 3014424..3016244 [-], 1821 | TraD | TraD_F, T4SS component |
| 58 | VCD_003714 | 3016293..3018443 [-], 2151 | TraI | Relaxase, MOBH Family |
| 59 | VCD_003715 | 3018598..3021249 [-], 2652 | hypothetical protein | |
| 60 | VCD_003716 | 3021293..3023368 [-], 2076 | hypothetical protein | |
| 61 | VCD_003717 | 3023385..3026042 [-], 2658 | hypothetical protein | |
| 62 | VCD_003718 | 3026042..3029668 [-], 3627 | hypothetical protein | |
| 63 | VCD_003719 | 3029684..3031447 [-], 1764 | hypothetical protein | |
| 64 | VCD_003720 | 3031464..3035141 [-], 3678 | hypothetical protein | |
| 65 | VCD_003721 | 3035160..3035753 [-], 594 | hypothetical protein | |
| 66 | VCD_003722 | 3035741..3036340 [-], 600 | hypothetical protein | |
| 67 | VCD_003723 | 3036350..3037276 [-], 927 | hypothetical protein | |
| 68 | VCD_003724 | 3037345..3037659 [-], 315 | hypothetical protein | |
| 69 | VCD_003725 | 3037747..3038652 [-], 906 | DNA polymerase III epsilon subunit-like 3'-5' exonuclease | |
| 70 | VCD_003726 | 3039291..3039740 [+], 450 | error-prone repair protein UmuD | |
| 71 | VCD_003727 | 3039709..3040017 [+], 309 | error-prone repair protein UmuC | |
| 72 | VCD_003728 | 3040602..3041801 [-], 1200 | transposase | |
| 73 | VCD_003729 | 3041840..3041947 [+], 108 | hypothetical protein | |
| 74 | VCD_003730 | 3042446..3043261 [+], 816 | dihydropteroate synthase | AR |
| 75 | VCD_003731 | 3043322..3044125 [+], 804 | aminoglycoside 3'-phosphotransferase | AR |
| 76 | VCD_003732 | 3044116..3044961 [+], 846 | aminoglycoside/hydroxyurea antibiotic resistance kinase | AR |
| 77 | VCD_003733 | 3045239..3045385 [+], 147 | hypothetical protein | |
| 78 | VCD_003734 | 3045633..3046655 [-], 1023 | cobalt-zinc-cadmium resistance protein czcD | |
| 79 | VCD_003735 | 3046790..3047173 [-], 384 | hypothetical protein | |
| 80 | VCD_003736 | 3047352..3047546 [+], 195 | StrA aminoglycoside phosphotransferase | |
| 81 | VCD_003737 | 3047684..3048361 [-], 678 | tetracycline repressor protein | |
| 82 | VCD_003738 | 3048365..3049639 [+], 1275 | TetA | AR |
| 83 | VCD_003739 | 3049891..3050196 [-], 306 | transcriptional regulator LysR family | |
| 84 | VCD_003740 | 3050224..3051438 [-], 1215 | florfenicol/chloramphenicol resistance protein | AR |
| 85 | VCD_003741 | 3051655..3052539 [-], 885 | hypothetical protein | |
| 86 | VCD_003742 | 3052570..3054063 [-], 1494 | IS phage Tn transposon-related protein | |
| 87 | VCD_003743 | 3054481..3056619 [-], 2139 | transposase Tn3 family protein | |
| 88 | VCD_003744 | 3057324..3057431 [+], 108 | hypothetical protein | |
| 89 | VCD_003745 | 3058519..3059622 [+], 1104 | error-prone repair protein UmuC | |
| 90 | VCD_003746 | 3059633..3060076 [-], 444 | hypothetical protein | |
| 91 | VCD_003747 | 3060099..3060233 [-], 135 | hypothetical protein | |
| 92 | VCD_003748 | 3060541..3061515 [+], 975 | Rod shape determination protein | |
| 93 | VCD_003749 | 3061512..3061787 [+], 276 | hypothetical protein | |
| 94 | VCD_003750 | 3061789..3063030 [+], 1242 | phage Integrase | |
| 95 | VCD_003751 | 3063160..3065178 [-], 2019 | hypothetical protein | |
| 96 | VCD_003752 | 3065951..3066049 [-], 99 | hypothetical protein | |
| 97 | VCD_003753 | 3066253..3068310 [+], 2058 | C-di-GMP phosphodiesterase A-related protein | |
| 98 | VCD_003754 | 3068299..3068772 [-], 474 | ribosomal-protein-S18p-alanine acetyltransferase | |
| 99 | VCD_003755 | 3068745..3069152 [-], 408 | DNA polymerase III psi subunit | |
| 100 | VCD_003756 | 3069263..3069814 [-], 552 | acetyltransferase | |
| 101 | VCD_003757 | 3069750..3070181 [-], 432 | hypothetical protein | |