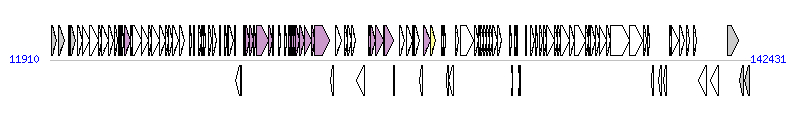The graph information of ICEYe1 components from FN298493 |
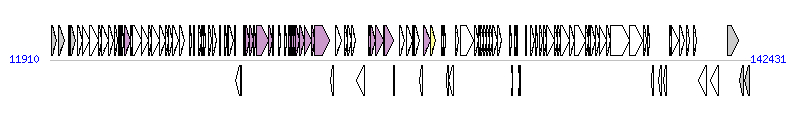 |
| Complete gene list of ICEYe1 from FN298493 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | Y69_0201 | 12248..13078 [+], 831 | putative integrase | |
| 2 | Y69_0200 | 13472..14704 [+], 1233 | putative helicase | |
| 3 | Y69_0199 | 15312..15620 [+], 309 | putative exported protein | |
| 4 | Y69_0198 | 15726..16688 [+], 963 | putative phage integrase | |
| 5 | Y69_0197 | 17037..17918 [+], 882 | conserved hypothetical protein | |
| 6 | dnaB | 17911..19278 [+], 1368 | DNA helicase | |
| 7 | Y69_0192 | 19278..20865 [+], 1588 | conserved hypothetical protein (pseudogene) | |
| 8 | Y69_0191 | 20862..21431 [+], 570 | conserved hypothetical protein | |
| 9 | Y69_0189 | 21537..22739 [+], 1203 | conserved hypothetical protein | |
| 10 | Y69_0187 | 22908..23666 [+], 759 | conserved hypothetical protein | |
| 11 | Y69_0185 | 23957..24499 [+], 543 | conserved hypothetical protein | |
| 12 | Y69_0184a | 24642..24884 [+], 243 | conserved hypothetical protein | |
| 13 | Y69_0184 | 24868..25116 [+], 249 | conserved hypothetical protein | |
| 14 | Y69_0183 | 25228..25659 [+], 432 | conserved hypothetical protein | |
| 15 | Y69_0181 | 25854..26828 [+], 975 | putative Type IV pilus lipoprotein (PilL) | Tfc2, T4SS component |
| 16 | Y69_0180 | 26830..27267 [+], 438 | putative Type IV pilus exported protein (PilM) | |
| 17 | Y69_0177 | 27279..28967 [+], 1689 | putative Type IV pilus lipoprotein (PilN) | |
| 18 | Y69_0176 | 28970..30277 [+], 1308 | putative Type IV pilus protein (PilO) | |
| 19 | Y69_0175 | 30264..30743 [+], 480 | putative Type IV pilus assembly protein (PilP) | |
| 20 | Y69_0169 | 30752..32305 [+], 1554 | putative Type IV pilus protein (PilQ) | |
| 21 | Y69_0168 | 32298..33407 [+], 1110 | putative Type IV pilus integral membrane protein (PilR) | |
| 22 | Y69_0167 | 33422..34009 [+], 588 | putative Type IV prepilin (PilS) | |
| 23 | Y69_0166 | 34006..34674 [+], 669 | putative prepilin peptidase (PilU) | |
| 24 | Y69_0164 | 34678..35955 [+], 1278 | putative prepilin (PilV) | |
| 25 | Y69_0163 | 36049..36981 [+], 933 | putative transposase | |
| 26 | Y69_0162 | 37857..38261 [+], 405 | conserved hypothetical protein | |
| 27 | Y69_0160 | 38576..38815 [+], 240 | putative transposase (pseudogene) | |
| 28 | Y69_0159 | 39606..40166 [+], 561 | hypothetical protein | |
| 29 | Y69_0158 | 40236..40508 [+], 273 | putative IS element protein (pseudogene) | |
| 30 | Y69_0156 | 40535..40929 [+], 395 | putative transposase (pseudogene) | |
| 31 | Y69_0155 | 41405..41983 [+], 579 | hypothetical protein | |
| 32 | Y69_0153 | 42241..42879 [+], 639 | hypothetical protein | |
| 33 | Y69_0152a | 43473..43688 [+], 216 | putative transposase (pseudogene) | |
| 34 | Y69_0152 | 44503..44835 [+], 333 | hypothetical protein | |
| 35 | Y69_0151 | 45038..45226 [+], 189 | hypothetical protein | |
| 36 | Y69_0150 | 45278..46096 [+], 819 | putative membrane protein | |
| 37 | Y69_0149 | 46241..46531 [+], 291 | putative antitoxin | |
| 38 | Y69_0147 | 46559..47353 [-], 795 | putative transposase (pseudogene) | |
| 39 | Y69_0146 | 47404..47694 [-], 291 | putative transposase | |
| 40 | Y69_0145 | 47907..48239 [+], 333 | conserved hypothetical protein | |
| 41 | Y69_0143 | 48331..49149 [+], 819 | putative membrane protein | Tfc3, T4SS component |
| 42 | Y69_0142 | 49128..49766 [+], 639 | putative exported lytic transglycosylase | Tfc4, T4SS component |
| 43 | Y69_0141 | 49763..50284 [+], 522 | putative exported protein | Tfc5, T4SS component |
| 44 | Y69_0138 | 50457..52577 [+], 2121 | conserved hypothetical protein | Tfc6, T4SS component |
| 45 | Y69_0137 | 52577..53329 [+], 753 | putative inner membrane protein | Tfc8, T4SS component |
| 46 | Y69_0136 | 53428..53543 [+], 116 | putative insertion element protein (pseudogene) | |
| 47 | Y69_0135 | 54422..54898 [+], 477 | hypothetical protein | |
| 48 | Y69_0134 | 55544..55981 [+], 438 | hypothetical protein | |
| 49 | Y69_0133 | 56437..56745 [+], 309 | putative exported protein | Tfc9, T4SS component |
| 50 | Y69_0131 | 56745..56990 [+], 246 | putative membrane protein | |
| 51 | Y69_0130 | 57006..57356 [+], 351 | putative membrane protein | Tfc10, T4SS component |
| 52 | Y69_0128 | 57366..57740 [+], 375 | putative membrane protein | Tfc11, T4SS component |
| 53 | Y69_0126 | 57737..58393 [+], 657 | putative membrane protein | Tfc12, T4SS component |
| 54 | Y69_0124 | 58393..59274 [+], 882 | conserved hypothetical protein | Tfc13, T4SS component |
| 55 | Y69_0121 | 59271..60740 [+], 1470 | putative exported protein | Tfc14, T4SS component |
| 56 | Y69_0120 | 60753..61169 [+], 417 | putative lipoprotein | Tfc15, T4SS component |
| 57 | Y69_0116 | 61169..63964 [+], 2796 | conserved hypothetical protein | Tfc16, T4SS component |
| 58 | Y69_0114 | 64191..64739 [-], 549 | conserved hypothetical protein | |
| 59 | Y69_0112 | 65111..66349 [+], 1239 | putative tranposase | |
| 60 | Y69_0111 | 66809..67459 [+], 651 | conserved hypothetical protein | |
| 61 | Y69_0110 | 67456..68172 [+], 717 | conserved hypothetical protein | |
| 62 | Y69_0109 | 68172..68825 [+], 654 | hypothetical protein | |
| 63 | Y69_0107 | 69149..70591 [-], 1443 | putative transposase | |
| 64 | Y69_0106 | 71279..71683 [+], 405 | putative exported protein | Tfc24, T4SS component |
| 65 | Y69_0105 | 71680..72642 [+], 963 | putative lipoprotein | Tfc23, T4SS component |
| 66 | Y69_0102 | 72654..74045 [+], 1392 | putative exported protein | Tfc22, T4SS component |
| 67 | Y69_0100 | 74045..74356 [+], 312 | putative outer membrane protein | |
| 68 | Y69_0098 | 74366..75886 [+], 1521 | putative membrane protein | Tfc19, T4SS component |
| 69 | Y69_0097 | 75921..76208 [-], 288 | conserved hypothetical protein | Tfc20, T4SS component |
| 70 | Y69_0095 | 77166..78053 [+], 888 | conserved hypothetical protein | |
| 71 | Y69_0094 | 78458..79459 [+], 1002 | conserved hypothetical protein | |
| 72 | Y69_0093 | 79456..79743 [+], 288 | conserved hypothetical protein | |
| 73 | Y69_0091 | 79844..80746 [+], 903 | conserved hypothetical protein | |
| 74 | Y69_0089 | 80811..81416 [-], 606 | putative membrane protein | |
| 75 | Y69_0087 | 81613..82833 [+], 1221 | putative helicase | Relaxase, MOBH Family |
| 76 | Y69_0086 | 82874..83830 [+], 957 | putative phage integrase | Integrase |
| 77 | Y69_0085 | 84907..85356 [+], 450 | putative transposase | |
| 78 | Y69_0084 | 85254..85571 [+], 318 | putative transposase (pseudogene) | |
| 79 | Y69_0083 | 85762..86241 [-], 480 | putative exported protein | |
| 80 | Y69_0081 | 86243..87172 [-], 930 | putative transcriptional regulator | |
| 81 | Y69_0078 | 87562..88101 [+], 540 | fimbria A protein | |
| 82 | Y69_0076 | 88428..90953 [+], 2526 | putative fimbrial usher protein | |
| 83 | Y69_0074 | 91063..91827 [+], 765 | putative fimbrial chaperone protein | |
| 84 | Y69_0072 | 91817..92152 [+], 336 | putative fimbrial membrane protein | |
| 85 | Y69_0070 | 92169..92666 [+], 498 | putative exported fimbrial protein | |
| 86 | Y69_0069 | 92793..93293 [+], 501 | putative exported fimbrial protein | |
| 87 | Y69_0068 | 93290..93796 [+], 507 | putative fimbrial membrane protein | |
| 88 | Y69_0064 | 93812..94351 [+], 540 | putative exported fimbrial protein | |
| 89 | Y69_0062 | 94344..95324 [+], 981 | putative reverse transcriptase | |
| 90 | Y69_0060 | 95327..96097 [+], 771 | hypothetical protein | |
| 91 | Y69_0058 | 97640..98023 [+], 384 | hypothetical protein | |
| 92 | Y69_0057a | 98115..98228 [-], 114 | putative transposase (pseudogene) | |
| 93 | Y69_0057 | 98504..98827 [+], 324 | conserved hypothetical protein | |
| 94 | Y69_0056b | 98814..99095 [+], 282 | putative DNA binding protein | |
| 95 | Y69_0056a | 99449..99517 [-], 69 | putative transposase (pseudogene) | |
| 96 | Y69_0056 | 99540..99716 [-], 177 | putative transposase (pseudogene) | |
| 97 | Y69_0054 | 100564..100791 [+], 228 | hypothetical protein | |
| 98 | Y69_0051 | 101511..102224 [+], 714 | putative exported protein | |
| 99 | Y69_0050 | 102519..102761 [+], 243 | hypothetical protein | |
| 100 | Y69_0049 | 103140..103772 [+], 633 | putative transcriptional regulator, GerE family | |
| 101 | Y69_0048 | 103988..104476 [+], 489 | conserved hypothetical protein | |
| 102 | Y69_0045 | 104473..105954 [+], 1482 | conserved hypothetical protein | |
| 103 | Y69_0044 | 105982..106473 [+], 492 | hypothetical protein | |
| 104 | Y69_0043 | 106586..107005 [+], 420 | conserved hypothetical protein | |
| 105 | Y69_0042 | 107017..108819 [+], 1803 | conserved hypothetical protein | |
| 106 | Y69_0041 | 108810..109757 [+], 948 | conserved hypothetical protein | |
| 107 | Y69_0040 | 109762..111816 [+], 2055 | conserved hypothetical protein | |
| 108 | Y69_0038 | 111824..112345 [+], 522 | hypothetical protein | |
| 109 | Y69_0037 | 112345..112644 [+], 300 | putative PAAR protein | |
| 110 | Y69_0034 | 112656..113699 [+], 1044 | ImpA-like protein | |
| 111 | Y69_0033 | 113696..114346 [+], 651 | putative lipoprotein | |
| 112 | Y69_0032 | 114346..115725 [+], 1380 | conserved hypothetical protein | |
| 113 | Y69_0031 | 115725..116339 [+], 615 | conserved hypothetical protein | |
| 114 | Y69_0027 | 116352..119888 [+], 3537 | putative outer membrane protein | |
| 115 | Y69_0022 | 119928..122471 [+], 2544 | putative Clp ATPase | |
| 116 | Y69_0021 | 122597..123223 [+], 627 | hypothetical protein | |
| 117 | Y69_0020 | 123370..123747 [+], 378 | hypothetical protein | |
| 118 | Y69_0019 | 124025..124447 [-], 423 | hypothetical protein | |
| 119 | Y69_0018 | 125397..125948 [-], 552 | hypothetical protein | |
| 120 | Y69_0017 | 126391..126855 [-], 465 | hypothetical protein | |
| 121 | Y69_0016 | 127355..127750 [+], 396 | putative exported protein | |
| 122 | Y69_0014 | 127753..129063 [+], 1311 | putative peptidyl-prolyl cis-trans isomerase | |
| 123 | Y69_0013 | 129311..130171 [+], 861 | putative hydrolase, alpha/beta fold family | |
| 124 | Y69_0012 | 130518..131135 [+], 618 | putative transcriptional regulator-GerE/LuxR family | |
| 125 | Y69_0011 | 131847..132431 [+], 585 | hypothetical protein | |
| 126 | Y69_0009 | 132789..134402 [-], 1614 | hypothetical protein | |
| 127 | Y69_0007 | 135055..136533 [-], 1479 | putative major facilitator superfamily (MFS) transporter | |
| 128 | speF | 138166..140328 [+], 2163 | ornithine decarboxylase | |
| 129 | Y69_0004 | 140397..141146 [-], 750 | putative membrane protein | |
| 130 | mltC | 141317..142393 [-], 1077 | membrane-bound lytic murein transglycosylase c precursor | |