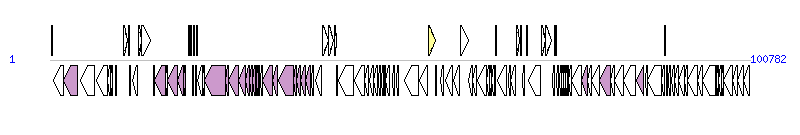The graph information of Pac_ICE3_cl components from KC148188 |
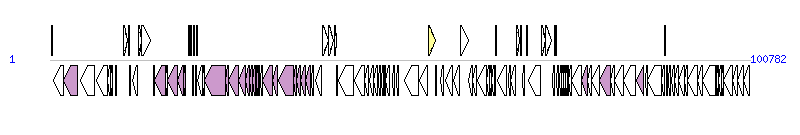 |
| Complete gene list of Pac_ICE3_cl from KC148188 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 268..393 [+], 126 | hypothetical protein | |
| 2 | - | 547..1962 [-], 1416 | integrase | |
| 3 | - | 1959..3899 [-], 1941 | pyruvate/2-oxoglutarate dehydrogenase complex dihydrolipoamide acyltransferase E2 component | Relaxase, MOBH Family |
| 4 | - | 4383..6368 [-], 1986 | methyl-accepting chemotaxis protein I | |
| 5 | - | 6706..8232 [-], 1527 | mobile element protein | |
| 6 | - | 8316..8681 [-], 366 | ISPpu14 transposase | |
| 7 | - | 8648..8980 [-], 333 | hypothetical protein | |
| 8 | - | 9430..9585 [-], 156 | hypothetical protein | |
| 9 | - | 10583..11176 [+], 594 | hypothetical protein | |
| 10 | - | 11311..11448 [+], 138 | hypothetical protein | |
| 11 | - | 11417..11542 [-], 126 | hypothetical protein | |
| 12 | - | 11918..12610 [-], 693 | protein of unknown function DUF159 | |
| 13 | - | 12721..13155 [+], 435 | error-prone repair protein UmuD | |
| 14 | - | 13142..14422 [+], 1281 | error-prone lesion bypass DNA polymerase V | |
| 15 | - | 14871..15020 [-], 150 | hypothetical protein | |
| 16 | - | 15043..16575 [-], 1533 | putative membrane protein | Tfc19, T4SS component |
| 17 | - | 16572..16892 [-], 321 | hypothetical protein | |
| 18 | - | 16949..18316 [-], 1368 | hypothetical protein | Tfc22, T4SS component |
| 19 | - | 18337..19278 [-], 942 | hypothetical protein | Tfc23, T4SS component |
| 20 | - | 19275..19562 [-], 288 | protein of unknown function DUF1525 | Tfc24, T4SS component |
| 21 | - | 19916..20104 [+], 189 | hypothetical protein | |
| 22 | - | 20162..20425 [+], 264 | hypothetical protein | |
| 23 | - | 20540..20692 [-], 153 | hypothetical protein | |
| 24 | - | 20648..20776 [+], 129 | hypothetical protein | |
| 25 | - | 20893..21063 [-], 171 | hypothetical protein | |
| 26 | - | 21101..21220 [+], 120 | hypothetical protein | |
| 27 | - | 21266..21961 [-], 696 | protein-disulfide isomerase | |
| 28 | - | 21958..22257 [-], 300 | hypothetical protein | |
| 29 | - | 22254..25271 [-], 3018 | type IV secretory pathway VirB4 component | Tfc16, T4SS component |
| 30 | - | 25271..25720 [-], 450 | putative lipoprotein | Tfc15, T4SS component |
| 31 | - | 25704..27191 [-], 1488 | putative exported protein | Tfc14, T4SS component |
| 32 | - | 27181..28104 [-], 924 | hypothetical protein | Tfc13, T4SS component |
| 33 | - | 28101..28769 [-], 669 | putative exported protein | Tfc12, T4SS component |
| 34 | - | 28766..29158 [-], 393 | hypothetical protein | Tfc11, T4SS component |
| 35 | - | 29174..29539 [-], 366 | hypothetical protein | Tfc10, T4SS component |
| 36 | - | 29559..29798 [-], 240 | hypothetical protein | |
| 37 | - | 29795..30016 [-], 222 | putative type III effector hop protein | Tfc9, T4SS component |
| 38 | - | 30036..30194 [-], 159 | hypothetical protein | |
| 39 | - | 30237..30569 [-], 333 | aconitase B | |
| 40 | - | 30579..32042 [-], 1464 | putative DNA helicase | TraI_F, T4SS component |
| 41 | - | 32052..32801 [-], 750 | putative membrane protein | Tfc8, T4SS component |
| 42 | - | 32838..34997 [-], 2160 | type IV secretory pathway VirD4 component | Tfc6, T4SS component |
| 43 | - | 35009..35515 [-], 507 | hypothetical protein | Tfc5, T4SS component |
| 44 | - | 35512..36069 [-], 558 | soluble lytic murein transglycosylase-related regulatory protein | Tfc4, T4SS component |
| 45 | - | 36054..36800 [-], 747 | putative exported protein STY4558 | Tfc3, T4SS component |
| 46 | - | 36809..37510 [-], 702 | methyl-accepting chemotaxis protein | Tfc2, T4SS component |
| 47 | - | 37602..37997 [-], 396 | hypothetical protein | |
| 48 | - | 38219..39142 [-], 924 | transcriptional regulator AraC family | |
| 49 | - | 39300..40058 [+], 759 | oxidoreductase short chain dehydrogenase/reductase family | |
| 50 | - | 40109..40819 [+], 711 | glutathione S-transferase | |
| 51 | - | 40900..41286 [+], 387 | hypothetical protein | |
| 52 | - | 41287..41424 [-], 138 | hypothetical protein | |
| 53 | - | 41478..43736 [-], 2259 | superfamily II DNA/RNA helicases SNF2 family | |
| 54 | - | 43833..45293 [-], 1461 | plasmid-related protein | |
| 55 | - | 45296..45922 [-], 627 | hypothetical protein | |
| 56 | - | 45988..46512 [-], 525 | hypothetical protein | |
| 57 | - | 46598..47209 [-], 612 | hypothetical protein | |
| 58 | - | 47311..47646 [-], 336 | hypothetical protein | |
| 59 | - | 47747..48049 [-], 303 | hypothetical protein | |
| 60 | - | 48126..48281 [-], 156 | hypothetical protein | |
| 61 | - | 48352..48906 [-], 555 | periplasmic protein TonB | |
| 62 | - | 49271..49636 [-], 366 | hypothetical protein | |
| 63 | - | 49926..50195 [-], 270 | hypothetical protein | |
| 64 | - | 51000..53021 [-], 2022 | DNA topoisomerase III | |
| 65 | - | 53240..54334 [-], 1095 | hypothetical protein | |
| 66 | - | 54536..55498 [+], 963 | SAM-like phage integrase | Integrase |
| 67 | - | 55472..55717 [-], 246 | hypothetical protein | |
| 68 | - | 56213..56593 [-], 381 | putative lipoprotein | |
| 69 | - | 56923..57915 [-], 993 | surface antigen | |
| 70 | - | 58073..58969 [-], 897 | LysR family transcriptional regulator YeiE | |
| 71 | - | 59114..60256 [+], 1143 | C4-dicarboxylate transporter/malic acid transport protein | |
| 72 | - | 60265..60867 [-], 603 | IS1415 transposase istA/B | |
| 73 | - | 60867..61412 [-], 546 | inorganic pyrophosphatase | |
| 74 | - | 61528..62811 [-], 1284 | enolase | |
| 75 | - | 62885..63226 [-], 342 | hypothetical protein | |
| 76 | - | 63321..63587 [-], 267 | hypothetical protein | |
| 77 | - | 63630..63953 [-], 324 | hypothetical protein | |
| 78 | - | 64034..64171 [-], 138 | hypothetical protein | |
| 79 | - | 64163..64294 [+], 132 | hypothetical protein | |
| 80 | - | 64396..65754 [-], 1359 | chloride channel protein | |
| 81 | - | 65991..66314 [-], 324 | hypothetical protein | |
| 82 | - | 66325..67068 [-], 744 | arsenical-resistance protein ACR3 | |
| 83 | - | 67160..67594 [+], 435 | transcriptional regulator MerR family | |
| 84 | - | 67744..67860 [+], 117 | hypothetical protein | |
| 85 | - | 68028..68252 [-], 225 | hypothetical protein | |
| 86 | - | 68565..68810 [+], 246 | hypothetical protein | |
| 87 | - | 68936..70555 [-], 1620 | methyl-accepting chemotaxis protein I | |
| 88 | - | 70802..71293 [+], 492 | hypothetical protein | |
| 89 | - | 71290..72249 [+], 960 | tyrosine recombinase XerC | |
| 90 | - | 72347..72694 [-], 348 | hypothetical protein | |
| 91 | - | 72647..72820 [+], 174 | hypothetical protein | |
| 92 | - | 72777..72956 [+], 180 | hypothetical protein | |
| 93 | - | 72920..73276 [-], 357 | hypothetical protein | |
| 94 | - | 73287..73607 [-], 321 | hypothetical protein | |
| 95 | - | 73607..73885 [-], 279 | hypothetical protein | |
| 96 | - | 73902..74177 [-], 276 | hypothetical protein | |
| 97 | - | 74251..74475 [-], 225 | hypothetical protein | |
| 98 | - | 74492..74650 [-], 159 | hypothetical protein | |
| 99 | - | 74651..75091 [-], 441 | conjugative transfer protein PilM | |
| 100 | - | 75122..76489 [-], 1368 | conjugative transfer pilus-tip adhesin protein PilV | |
| 101 | - | 76486..77418 [-], 933 | conjugative transfer ATPase PilU | TraJ_I, T4SS component |
| 102 | - | 77421..77942 [-], 522 | conjugative transfer protein PilS | |
| 103 | - | 77980..79071 [-], 1092 | conjugative transfer protein PilR | |
| 104 | - | 79061..80686 [-], 1626 | bundle-forming pilus protein BfpD | DotB, T4SS component |
| 105 | - | 80683..81234 [-], 552 | conjugative transfer protein PilP | |
| 106 | - | 81275..82633 [-], 1359 | conjugative transfer protein PilO | |
| 107 | - | 82635..84368 [-], 1734 | conjugative transfer protein PilN | |
| 108 | - | 84371..85513 [-], 1143 | conjugative transfer protein PilL | Tfc2, T4SS component |
| 109 | - | 85749..85997 [-], 249 | hypothetical protein | |
| 110 | - | 86096..88048 [-], 1953 | DNA/RNA helicase | |
| 111 | - | 88061..88414 [-], 354 | hypothetical protein | |
| 112 | - | 88482..88664 [+], 183 | hypothetical protein | |
| 113 | - | 88766..88966 [-], 201 | hypothetical protein | |
| 114 | - | 89101..89343 [-], 243 | hypothetical protein | |
| 115 | - | 89529..89978 [-], 450 | single-stranded DNA-binding protein | |
| 116 | - | 89980..90552 [-], 573 | integrase regulator R | |
| 117 | - | 90552..91331 [-], 780 | hypothetical protein | |
| 118 | - | 91324..91836 [-], 513 | haemin uptake system outer membrane receptor | |
| 119 | - | 91839..93209 [-], 1371 | hypothetical protein | |
| 120 | - | 93209..93946 [-], 738 | hypothetical protein | |
| 121 | - | 93983..95746 [-], 1764 | protein with parB-like nuclease domain | |
| 122 | - | 95749..95994 [-], 246 | alginate biosynthesis transcriptional activator | |
| 123 | - | 96001..96246 [-], 246 | hypothetical protein | |
| 124 | - | 96243..96644 [-], 402 | hypothetical protein | |
| 125 | - | 96646..96987 [-], 342 | hypothetical protein | |
| 126 | - | 96984..98330 [-], 1347 | replicative DNA helicase | |
| 127 | - | 98330..99028 [-], 699 | hypothetical protein | |
| 128 | - | 99025..99780 [-], 756 | hypothetical protein | |
| 129 | - | 99782..100642 [-], 861 | parA-like chromosome partitioning ATPase | |