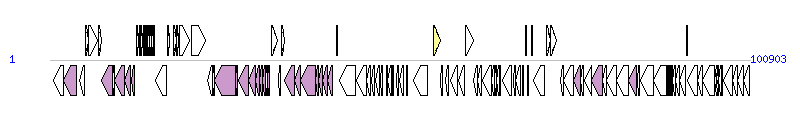The graph information of Pac_ICE1_nz components from KC148184 |
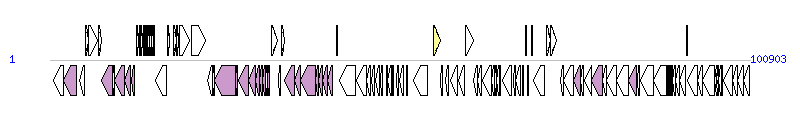 |
| Complete gene list of Pac_ICE1_nz from KC148184 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 549..1952 [-], 1404 | integrase | |
| 2 | - | 1949..3865 [-], 1917 | pyruvate/2-oxoglutarate dehydrogenase complex dihydrolipoamide acyltransferase E2 component | Relaxase, MOBH Family |
| 3 | - | 4282..4974 [-], 693 | protein of unknown function DUF159 | |
| 4 | - | 5088..5522 [+], 435 | error-prone repair protein UmuD | |
| 5 | - | 5509..6789 [+], 1281 | error-prone lesion bypass DNA polymerase V | |
| 6 | - | 7009..7398 [+], 390 | hypothetical protein | |
| 7 | - | 7449..8987 [-], 1539 | putative membrane protein | Tfc19, T4SS component |
| 8 | - | 8984..9358 [-], 375 | hypothetical protein | |
| 9 | - | 9361..10728 [-], 1368 | hypothetical protein | Tfc22, T4SS component |
| 10 | - | 10749..11555 [-], 807 | hypothetical protein | Tfc23, T4SS component |
| 11 | - | 11684..12115 [-], 432 | hypothetical protein | Tfc24, T4SS component |
| 12 | - | 12448..12699 [+], 252 | hypothetical protein | |
| 13 | - | 12846..12968 [+], 123 | hypothetical protein | |
| 14 | - | 13071..13334 [+], 264 | hypothetical protein | |
| 15 | - | 13469..13657 [+], 189 | hypothetical protein | |
| 16 | - | 13720..13962 [+], 243 | hypothetical protein | |
| 17 | - | 14053..14289 [+], 237 | hypothetical protein | |
| 18 | - | 14378..14602 [+], 225 | hypothetical protein | |
| 19 | - | 14656..14907 [+], 252 | hypothetical protein | |
| 20 | - | 14917..15132 [+], 216 | hypothetical protein | |
| 21 | - | 15182..16732 [-], 1551 | C-5 cytosine-specific DNA methylase | |
| 22 | - | 17006..17200 [+], 195 | hypothetical protein | |
| 23 | - | 17872..18204 [+], 333 | hypothetical protein | |
| 24 | - | 18201..18536 [+], 336 | ISPpu14 transposase | |
| 25 | - | 18620..20146 [+], 1527 | mobile element protein | |
| 26 | - | 20418..22469 [+], 2052 | methyl-accepting chemotaxis protein I | |
| 27 | - | 22774..23460 [-], 687 | protein-disulfide isomerase | |
| 28 | - | 23457..23756 [-], 300 | hypothetical protein | |
| 29 | - | 23753..26761 [-], 3009 | type IV secretory pathway VirB4 component | Tfc16, T4SS component |
| 30 | - | 26761..27081 [-], 321 | putative lipoprotein | Tfc15, T4SS component |
| 31 | - | 27194..28678 [-], 1485 | putative exported protein | Tfc14, T4SS component |
| 32 | - | 28668..29591 [-], 924 | hypothetical protein | Tfc13, T4SS component |
| 33 | - | 29588..30256 [-], 669 | putative exported protein | Tfc12, T4SS component |
| 34 | - | 30253..30645 [-], 393 | hypothetical protein | Tfc11, T4SS component |
| 35 | - | 30661..31029 [-], 369 | hypothetical protein | Tfc10, T4SS component |
| 36 | - | 31049..31288 [-], 240 | hypothetical protein | |
| 37 | - | 31285..31689 [-], 405 | putative type III effector hop protein | Tfc9, T4SS component |
| 38 | - | 31898..32845 [+], 948 | hypothetical protein | |
| 39 | - | 32903..33217 [-], 315 | aconitase B | |
| 40 | - | 33385..33816 [+], 432 | hypothetical protein | |
| 41 | - | 33840..35297 [-], 1458 | putative DNA helicase | TraI_F, T4SS component |
| 42 | - | 35307..36056 [-], 750 | putative membrane protein | Tfc8, T4SS component |
| 43 | - | 36096..38255 [-], 2160 | type IV secretory pathway VirD4 component | Tfc6, T4SS component |
| 44 | - | 38267..38773 [-], 507 | hypothetical protein | Tfc5, T4SS component |
| 45 | - | 38770..39327 [-], 558 | soluble lytic murein transglycosylase-related regulatory protein | Tfc4, T4SS component |
| 46 | - | 39312..40058 [-], 747 | putative exported protein STY4558 | Tfc3, T4SS component |
| 47 | - | 40067..40777 [-], 711 | methyl-accepting chemotaxis protein | Tfc2, T4SS component |
| 48 | - | 41327..41461 [+], 135 | hypothetical protein | |
| 49 | - | 41800..44064 [-], 2265 | superfamily II DNA/RNA helicases SNF2 family | |
| 50 | - | 44171..45622 [-], 1452 | plasmid-related protein | |
| 51 | - | 45652..46251 [-], 600 | hypothetical protein | |
| 52 | - | 46318..46842 [-], 525 | hypothetical protein | |
| 53 | - | 46931..47542 [-], 612 | hypothetical protein | |
| 54 | - | 47638..47973 [-], 336 | hypothetical protein | |
| 55 | - | 48450..48605 [-], 156 | hypothetical protein | |
| 56 | - | 48676..49248 [-], 573 | periplasmic protein TonB | |
| 57 | - | 49345..49575 [-], 231 | hypothetical protein | |
| 58 | - | 49880..50245 [-], 366 | hypothetical protein | |
| 59 | - | 50430..50978 [-], 549 | putative regulatory protein | |
| 60 | - | 51327..51593 [-], 267 | hypothetical protein | |
| 61 | - | 52413..54434 [-], 2022 | DNA topoisomerase III | |
| 62 | - | 55328..56290 [+], 963 | SAM-like phage integrase | Integrase |
| 63 | - | 56264..56509 [-], 246 | hypothetical protein | |
| 64 | - | 57005..57385 [-], 381 | putative lipoprotein | |
| 65 | - | 57715..58707 [-], 993 | surface antigen | |
| 66 | - | 58865..59761 [-], 897 | LysR family transcriptional regulator YeiE | |
| 67 | - | 59879..61048 [+], 1170 | C4-dicarboxylate transporter/malic acid transport protein | |
| 68 | - | 61057..61659 [-], 603 | IS1415 transposase istA/B | |
| 69 | - | 61659..62204 [-], 546 | inorganic pyrophosphatase | |
| 70 | - | 62320..63603 [-], 1284 | enolase | |
| 71 | - | 63677..64018 [-], 342 | hypothetical protein | |
| 72 | - | 64113..64379 [-], 267 | hypothetical protein | |
| 73 | - | 64422..64745 [-], 324 | hypothetical protein | |
| 74 | - | 64826..64963 [-], 138 | hypothetical protein | |
| 75 | - | 65188..66534 [-], 1347 | chloride channel protein | |
| 76 | - | 66783..67106 [-], 324 | hypothetical protein | |
| 77 | - | 67117..67860 [-], 744 | arsenical-resistance protein ACR3 | |
| 78 | - | 68087..68215 [-], 129 | hypothetical protein | |
| 79 | - | 68536..68652 [+], 117 | hypothetical protein | |
| 80 | - | 68820..69044 [-], 225 | hypothetical protein | |
| 81 | - | 69357..69602 [+], 246 | hypothetical protein | |
| 82 | - | 69728..71347 [-], 1620 | methyl-accepting chemotaxis protein I | |
| 83 | - | 71540..72100 [+], 561 | hypothetical protein | |
| 84 | - | 72085..73044 [+], 960 | tyrosine recombinase XerC | |
| 85 | - | 73603..74043 [-], 441 | conjugative transfer protein PilM | |
| 86 | - | 74075..75481 [-], 1407 | conjugative transfer pilus-tip adhesin protein PilV | |
| 87 | - | 75478..76410 [-], 933 | conjugative transfer ATPase PilU | TraJ_I, T4SS component |
| 88 | - | 76413..76937 [-], 525 | conjugative transfer protein PilS | |
| 89 | - | 76976..78067 [-], 1092 | conjugative transfer protein PilR | |
| 90 | - | 78057..79688 [-], 1632 | conjugative transfer ATPase PilQ | TraJ_I, T4SS component |
| 91 | - | 79685..80287 [-], 603 | conjugative transfer protein PilP | |
| 92 | - | 80277..81635 [-], 1359 | conjugative transfer protein PilO | |
| 93 | - | 81637..83367 [-], 1731 | conjugative transfer protein PilN | |
| 94 | - | 83370..84503 [-], 1134 | conjugative transfer protein PilL | Tfc2, T4SS component |
| 95 | - | 84708..84956 [-], 249 | hypothetical protein | |
| 96 | - | 85056..87008 [-], 1953 | DNA/RNA helicase | |
| 97 | - | 87011..88897 [-], 1887 | hypothetical protein | |
| 98 | - | 89002..89184 [-], 183 | hypothetical protein | |
| 99 | - | 89265..89510 [-], 246 | hypothetical protein | |
| 100 | - | 89564..89683 [-], 120 | hypothetical protein | |
| 101 | - | 89717..90187 [-], 471 | single-stranded DNA-binding protein | |
| 102 | - | 90189..90764 [-], 576 | integrase regulator R | |
| 103 | - | 90770..91549 [-], 780 | hypothetical protein | |
| 104 | - | 91692..91892 [+], 201 | hypothetical protein | |
| 105 | - | 91949..93313 [-], 1365 | hypothetical protein | |
| 106 | - | 93319..94056 [-], 738 | hypothetical protein | |
| 107 | - | 94093..95853 [-], 1761 | protein with parB-like nuclease domain | |
| 108 | - | 95856..96173 [-], 318 | alginate biosynthesis transcriptional activator | |
| 109 | - | 96109..96354 [-], 246 | hypothetical protein | |
| 110 | - | 96351..96758 [-], 408 | hypothetical protein | |
| 111 | - | 96760..96933 [-], 174 | hypothetical protein | |
| 112 | - | 97098..98429 [-], 1332 | replicative DNA helicase | |
| 113 | - | 98444..99145 [-], 702 | hypothetical protein | |
| 114 | - | 99142..99897 [-], 756 | hypothetical protein | |
| 115 | - | 99899..100759 [-], 861 | parA-like chromosome partitioning ATPase | |