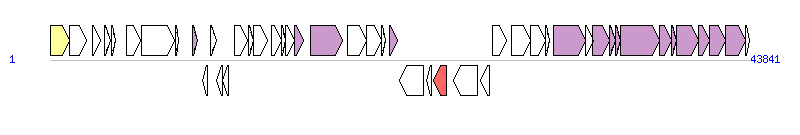The graph information of ICETn43716061 components from KP299160 |
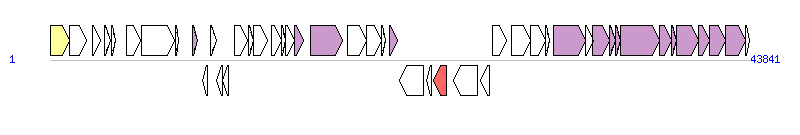 |
| Complete gene list of ICETn43716061 from KP299160 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | ICEPaeSP_00001 | 9..1208 [+], 1200 | integrase | Integrase |
| 2 | ICEPaeSP_00002 | 1205..2269 [+], 1065 | hypothetical protein | |
| 3 | ICEPaeSP_00003 | 2634..3143 [+], 510 | hypothetical protein | |
| 4 | ICEPaeSP_00004 | 3427..3777 [+], 351 | hypothetical protein | |
| 5 | ICEPaeSP_00005 | 3843..4121 [+], 279 | hypothetical protein | |
| 6 | ICEPaeSP_00006 | 4822..5646 [+], 825 | hypothetical protein | |
| 7 | spo0C | 5711..7768 [+], 2058 | Chromosome-partitioning protein Spo0J | |
| 8 | ICEPaeSP_00008 | 7830..8039 [+], 210 | hypothetical protein | |
| 9 | ICEPaeSP_00009 | 8894..9208 [+], 315 | hypothetical protein | TraF, T4SS component |
| 10 | ICEPaeSP_00010 | 9543..9836 [-], 294 | anaerobic benzoate catabolism transcriptional regulator | |
| 11 | ICEPaeSP_00011 | 10060..10407 [+], 348 | hypothetical protein | |
| 12 | ICEPaeSP_00012 | 10414..10803 [-], 390 | Transposase DDE domain protein | |
| 13 | ICEPaeSP_00013 | 10773..11195 [-], 423 | hypothetical protein | |
| 14 | ICEPaeSP_00014 | 11577..12356 [+], 780 | hypothetical protein | |
| 15 | ICEPaeSP_00015 | 12440..12721 [+], 282 | Helix-turn-helix domain protein | |
| 16 | ICEPaeSP_00016 | 12748..13605 [+], 858 | Replication initiator protein A | |
| 17 | soj | 13859..14497 [+], 639 | Chromosome-partitioning ATPase Soj | |
| 18 | ICEPaeSP_00018 | 14494..14778 [+], 285 | hypothetical protein | |
| 19 | ICEPaeSP_00019 | 14775..15308 [+], 534 | hypothetical protein | |
| 20 | ICEPaeSP_00020 | 15305..15880 [+], 576 | Peptidase S26 | TraF, T4SS component |
| 21 | ICEPaeSP_00021 | 16329..18302 [+], 1974 | hypothetical protein | Relaxase, MOBL Family |
| 22 | bcr_1 | 18610..19797 [+], 1188 | Bicyclomycin resistance protein | |
| 23 | catM_1 | 19807..20751 [+], 945 | HTH-type transcriptional regulator CatM | |
| 24 | ICEPaeSP_00024 | 20748..21014 [+], 267 | hypothetical protein | |
| 25 | ICEPaeSP_00025 | 21250..21738 [+], 489 | hypothetical protein | TraG, T4SS component |
| 26 | ICEPaeSP_00026 | 21881..23404 [-], 1524 | Putative transposase | |
| 27 | groL_1 | 23592..23894 [-], 303 | 60 kDa chaperonin | |
| 28 | ICEPaeSP_00028 | 24005..24835 [-], 831 | Beta-lactamase IMP-1 precursor | AR |
| 29 | ICEPaeSP_00029 | 25255..26778 [-], 1524 | Putative transposase | |
| 30 | groL_2 | 26966..27502 [-], 537 | 60 kDa chaperonin | |
| 31 | ICEPaeSP_00031 | 27697..28602 [+], 906 | hypothetical protein | |
| 32 | bcr_2 | 28910..30097 [+], 1188 | Bicyclomycin resistance protein | |
| 33 | catM_2 | 30107..31051 [+], 945 | HTH-type transcriptional regulator CatM | |
| 34 | ICEPaeSP_00034 | 31048..31314 [+], 267 | hypothetical protein | |
| 35 | traG | 31550..33550 [+], 2001 | Conjugal transfer protein TraG | VirD4, T4SS component |
| 36 | ICEPaeSP_00036 | 33547..34011 [+], 465 | hypothetical protein | |
| 37 | ptlH | 34008..35060 [+], 1053 | Type IV secretion system protein PtlH | TrbB, T4SS component |
| 38 | ICEPaeSP_00038 | 35057..35440 [+], 384 | TrbC/VIRB2 family protein | TrbC, T4SS component |
| 39 | ICEPaeSP_00039 | 35437..35709 [+], 273 | Type IV secretory pathway, VirB3-like protein | TrbD, T4SS component |
| 40 | virB4 | 35722..38187 [+], 2466 | Type IV secretion system protein virB4 | TrbE, T4SS component |
| 41 | ICEPaeSP_00041 | 38184..38939 [+], 756 | hypothetical protein | TrbJ, T4SS component |
| 42 | ICEPaeSP_00042 | 38951..39250 [+], 300 | hypothetical protein | TrbJ, T4SS component |
| 43 | ICEPaeSP_00043 | 39247..40596 [+], 1350 | TrbL/VirB6 plasmid conjugal transfer protein | TrbL, T4SS component |
| 44 | ICEPaeSP_00044 | 40608..41312 [+], 705 | VirB8 protein | TrbF, T4SS component |
| 45 | ptlF | 41309..42301 [+], 993 | Type IV secretion system protein PtlF precursor | TrbG, T4SS component |
| 46 | ICEPaeSP_00046 | 42304..43587 [+], 1284 | Type IV secretion system protein virB10 | TrbI, T4SS component |
| 47 | ICEPaeSP_00047 | 43584..43829 [+], 246 | hypothetical protein | |