
| Integrative and conjugative element (ICE) |
Integrative and conjugative element (ICE) is an important member of the bacterial mobile genetic elements,
which is integrative to the bacterial chromosome and encodes a fully functioning conjugation machinery
and is thus self-transmissible between bacterial cells. The cargo genes of ICEs, coding for the antibiotic
resistance determinants and the virulence factors, can confer the host with selective advantages, making
the ICE a vital driving force for bacterial adaptation and evolution. In addition to the self-transferability,the ICEs have also been reported capable to mobilize other genetic elements, such as the chromosome-borne integrative and mobilizable elements (IMEs), cis-mobilizable elements (CIMEs), plasmids. IMEs and CIMEs are also important vehicles for the spread of antiobiotic resistances and virulence factors. However, unlike ICE, IMEs and CIMEs are commonly devoid of conjugal apparatus, thus have to hijack the machinery of other conjugative elements. The crosstalk between IMEs, CIMEs and ICEs is an emerging hotspot in the research of horizontal gene transfer (Bellanger, et al., FEMS Microbiol Rev, 2014; Delavat, et al., FEMS Microbiol Rev, 2017; Guédon, et al., Genes, 2017). |
| ICEfinder: a web-based tool for the detection of ICEs/IMEs of bacterial genomes |
|
ICEfinder was designed to facilitate rapid detection of T4SS-type ICEs, AICEs and IMEs in bacterial genome sequences.
ICEfinder first detects the signature sequences of the recombination modules and conjugation modules based on the profile HMMs.
It also searches for the oriT region using the approach proposed by oriTfinder
(Li, et al., Nucleic Acids Res, 2018).
Then it co-localizes, filters and groups the corresponding genes (Figure 1). Finally, those elements carrying an integrase gene,
a relaxase gene, and T4SS gene clusters (Li, et al., Brief Bioinform, 2018)
are considered as ICEs, while those without T4SS but with integrase and relaxase are tagged as putative IMEs.
ICEfinder also tries to detect some particular IMEs with integrase and an oriT but no relaxase.
For those without T4SS but with integrase, replication and the
AICE translocation-related proteins are thought to be putative AICEs. ICEfinder employs ARAGORN
(Laslett, et al., Nucleic Acids Res, 2004)
with the default parameters to identify the 3’ termini of the tRNA/tmRNA genes as the putative
ICE insertion sites and Vmatch with the default
options to detect the directed repeats (DR) as the tRNA-distal boundaries. The ICE-coding putative
acquired antibiotic resistance genes (ARG) and virulence factors (VF) are also identified by NCBI BLASTp
(Camacho, et al., BMC Bioinformatics, 2009)
with the cutoff of Ha-value of 0.64.
|
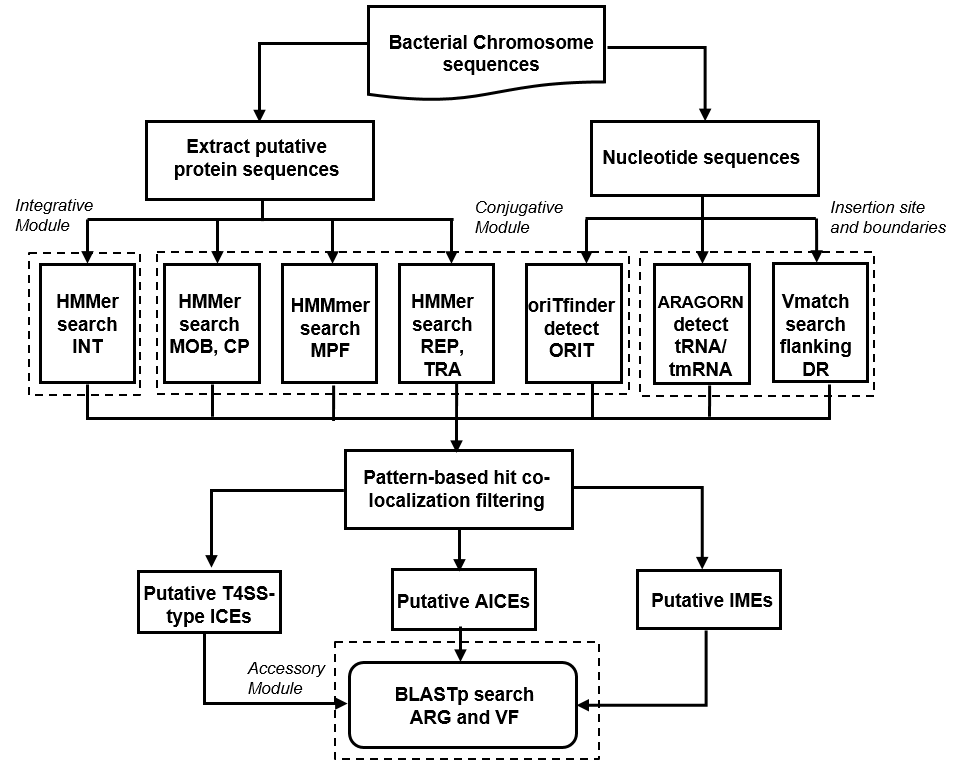 Figure 1. The prediction strategy used by ICEfinder to identify ICEs/IMEs of bacterial genomes. |
| Detection the putative virulence factors (VFs) and acquired antibiotic resistance determinants (ARs) with the protein sequence similarity by using BLASTp-based Ha-value. |
|
To examine the degree of sequence similarities at an amino acid level between each query protein and the oriTfinder-collected
VFs and ARs, the NCBI BLASTp-derived Ha-value was employed. For each query, the Ha-value was calculated as follows:
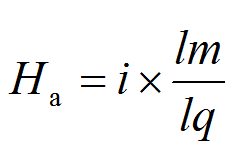 |