Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103257 |
| Name | oriT_pPg2708 |
| Organism | Pseudomonas coronafaciens pv. garcae strain 2708 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KY362367 (67391..67559 [+], 169 nt) |
| oriT length | 169 nt |
| IRs (inverted repeats) | 125..132, 136..143 (CAAAGGGG..CCCCTTTG) 87..93, 102..108 (CAAAAAG..CTTTTTG) 32..38, 40..46 (TTGGGTC..GACCCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 169 nt
>oriT_pPg2708
CTGTGTCACCGAGAGACATCCAGAGACAATCTTGGGTCTGACCCAAAATCTCGAATTCGCAAAACAGGCACGTTGACGATTGTTTACAAAAAGAGCTTCGCCTTTTTGAAAACATCGTCCGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAGAGCGTCG
CTGTGTCACCGAGAGACATCCAGAGACAATCTTGGGTCTGACCCAAAATCTCGAATTCGCAAAACAGGCACGTTGACGATTGTTTACAAAAAGAGCTTCGCCTTTTTGAAAACATCGTCCGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAGAGCGTCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file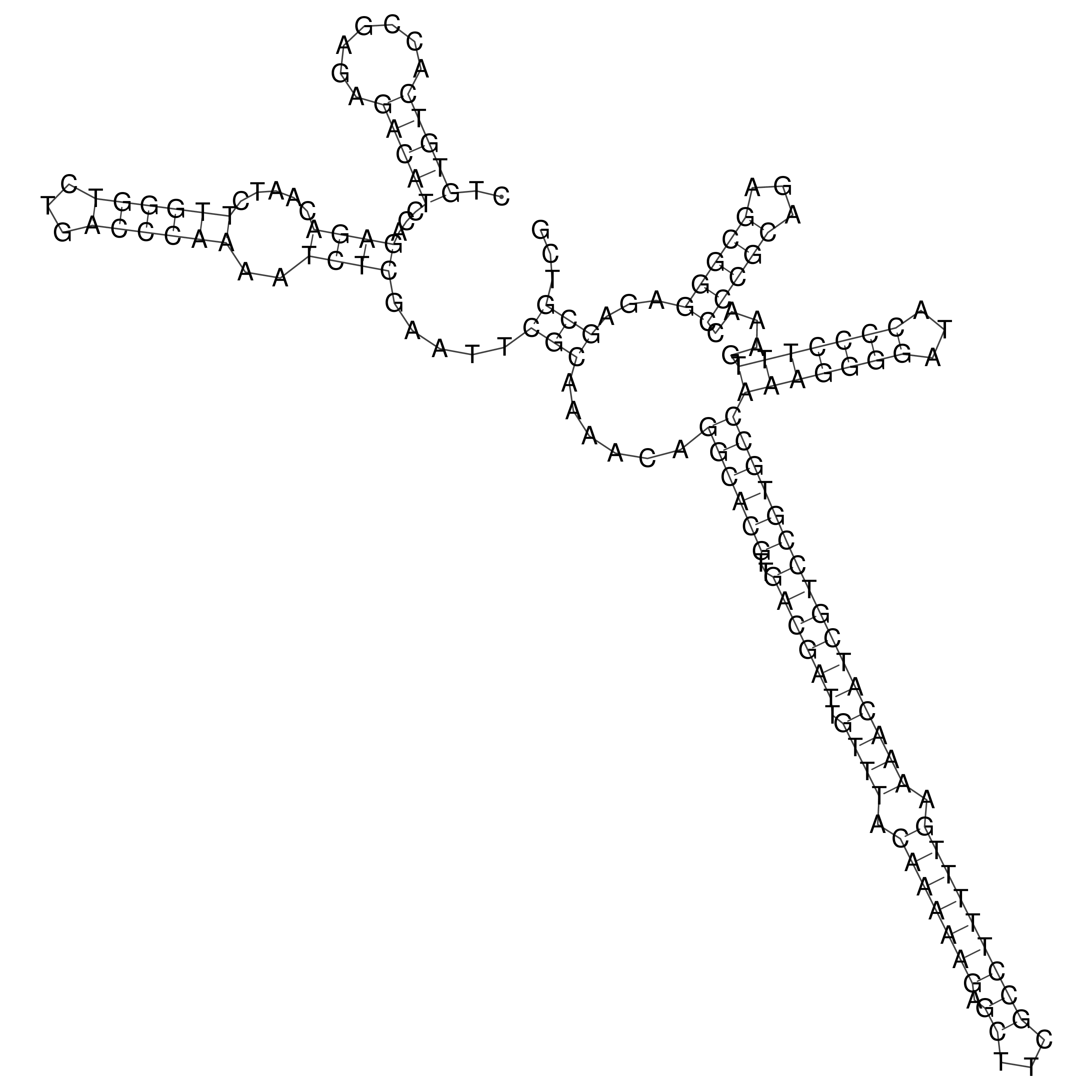
Relaxase
| ID | 2434 | GenBank | WP_235671735 |
| Name | Relaxase_HTL68_RS00450_pPg2708 |
UniProt ID | _ |
| Length | 1257 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1257 a.a. Molecular weight: 139229.05 Da Isoelectric Point: 9.4299
>WP_235671735.1 LPD7 domain-containing protein [Pseudomonas coronafaciens]
MLIRVRGYNAGVQEYLEEGIKSGREFTRDELDHRVILYGDLDLTRMVYESIEDKGQDRYLTITLAFREDH
IPHAMLDSITQEFKQFMMYAYKEEEFNFYAEAHLPKIKTITDKKTGEPIDRKPHIHIVIPKKNLLSGNIF
DPVGSNYAQSEKHLEAFQEYVNNKYNLSSPREHVRADIKDAASVLSRYKGDDFYGKNRQFKQDLVKQVIE
QGIQTRSGFYALVGEHGETRVRNKDKANEYIAVKLPGDAKFTNLKDTIFHDDFIVRRELKKPPLDQKIIS
DRLLAWPQRAKEIKYISKAGPDFRKLYVASSSEERVRILNECEQKFYQTYGESHDSVHTGQRQRDHQRSS
AETAGRRTAEPADGLQDLSVSNVADHRQAGSARSRDGALLLPSDAHVHLGQSQPGGDSGLRPSVPGGGRG
RRNESTTERGRGGSGPAAVSQEATGTATPAGATGRRRTGAGKPRNARVRAGRLIPPYAKNPHRVATIADI
EERGRRLFDPLKKPSDNALVFYRAPSVTSMPKAATATAPGRSTAGRRPRTSGKPRPPRQWRPGSVVPPYA
KNPHRVATVADIEQRARMLFEPLKRPADKALVFKRASIKALTVNRQASTVAAYFTREAQHNRIAPAHRQA
IRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHKPQPNTEVNFMGSAAIRNLLEDEKE
DPGFSISGARGPGPAGVRDQVKRVMDRFAKQIDPVAASQRARDLSAKDLYTRKAKFSQNVHYLDKQTDKT
LFVDTGKSISMRRTGITESGVSVALQLAKERFGSTLTITGTAEFKKLVIEAVAKNGLDVHFTDKAMNQSL
ADRRAELDIERDGLSIGPANDLARHVDDSTRDVRDQADRMGVTVRIEALYGEGKTAEQVSQALAVQLDTV
PESERIAFVEKVAITLGIPERGEPKGDQAFAQWQAQRAQPAADSEATARSEAQADVSTPSDTAPEAVTPT
AATNSASASPEAAKPARVNDPDLQSPSELVRLEAQWRRDFPMSEADVRASDTVMALRGEDHAIWIIATDD
KTPEAAAMLASYMENDSYREAFKSSIVAAYKTVENSPQSVDDLDHLTAMAAQIVNEVEDRLYSAPQAASG
QATPSSSKVIEGALIEHGEAPYQHKKDNQMSYFVTLKPEGGKPRTVWGVGLEYAINESGLKPGDQVRLKD
LGTRPVVVQVIAEDGTTTDKTVNRREWSAQPTAPEREVAETTPKGQAIAAGTPELSSPDEDDGMSVD
MLIRVRGYNAGVQEYLEEGIKSGREFTRDELDHRVILYGDLDLTRMVYESIEDKGQDRYLTITLAFREDH
IPHAMLDSITQEFKQFMMYAYKEEEFNFYAEAHLPKIKTITDKKTGEPIDRKPHIHIVIPKKNLLSGNIF
DPVGSNYAQSEKHLEAFQEYVNNKYNLSSPREHVRADIKDAASVLSRYKGDDFYGKNRQFKQDLVKQVIE
QGIQTRSGFYALVGEHGETRVRNKDKANEYIAVKLPGDAKFTNLKDTIFHDDFIVRRELKKPPLDQKIIS
DRLLAWPQRAKEIKYISKAGPDFRKLYVASSSEERVRILNECEQKFYQTYGESHDSVHTGQRQRDHQRSS
AETAGRRTAEPADGLQDLSVSNVADHRQAGSARSRDGALLLPSDAHVHLGQSQPGGDSGLRPSVPGGGRG
RRNESTTERGRGGSGPAAVSQEATGTATPAGATGRRRTGAGKPRNARVRAGRLIPPYAKNPHRVATIADI
EERGRRLFDPLKKPSDNALVFYRAPSVTSMPKAATATAPGRSTAGRRPRTSGKPRPPRQWRPGSVVPPYA
KNPHRVATVADIEQRARMLFEPLKRPADKALVFKRASIKALTVNRQASTVAAYFTREAQHNRIAPAHRQA
IRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHKPQPNTEVNFMGSAAIRNLLEDEKE
DPGFSISGARGPGPAGVRDQVKRVMDRFAKQIDPVAASQRARDLSAKDLYTRKAKFSQNVHYLDKQTDKT
LFVDTGKSISMRRTGITESGVSVALQLAKERFGSTLTITGTAEFKKLVIEAVAKNGLDVHFTDKAMNQSL
ADRRAELDIERDGLSIGPANDLARHVDDSTRDVRDQADRMGVTVRIEALYGEGKTAEQVSQALAVQLDTV
PESERIAFVEKVAITLGIPERGEPKGDQAFAQWQAQRAQPAADSEATARSEAQADVSTPSDTAPEAVTPT
AATNSASASPEAAKPARVNDPDLQSPSELVRLEAQWRRDFPMSEADVRASDTVMALRGEDHAIWIIATDD
KTPEAAAMLASYMENDSYREAFKSSIVAAYKTVENSPQSVDDLDHLTAMAAQIVNEVEDRLYSAPQAASG
QATPSSSKVIEGALIEHGEAPYQHKKDNQMSYFVTLKPEGGKPRTVWGVGLEYAINESGLKPGDQVRLKD
LGTRPVVVQVIAEDGTTTDKTVNRREWSAQPTAPEREVAETTPKGQAIAAGTPELSSPDEDDGMSVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2201 | GenBank | WP_122326400 |
| Name | t4cp2_HTL68_RS00365_pPg2708 |
UniProt ID | _ |
| Length | 553 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 553 a.a. Molecular weight: 61808.33 Da Isoelectric Point: 6.1516
>WP_122326400.1 type IV secretory system conjugative DNA transfer family protein [Pseudomonas coronafaciens]
MARVKSKPISPDNPQERIDEHPAFLLGKHPTEDSFLASYGQQFVMLAAPPGTGKGVGAVIPNLLSYPDSM
VVNDPKFENWEITSGFRASTGHKVYRFSPERLETHRWNPLSAINRDPLYRLGEIRTIARVLFVSDNPKNQ
EWYNKAGNVFTAMLLYLMETPEMPCTLPQAYEIGSLGTGIGTWAQQIIQLRSSGPNALSFETLRELNGVY
EASKNKSSGWSTTVDIVRDVLSVYAEKTVAWAVSGDDIDLSKAREEKMTAYFSVTEGSLKKYGPLMNLFF
TQAIRLNSKVIPEQGGHCADGTLRYKYQLALLMDELAIMGRIESLETAPALTRGAGLRFFFIFQGKDQLR
AIYGEEAANAIMKAIHNEIVFAPGDIKLAEEYSRRLGNTTVRVHNQSLNRQKHEVGARGQTDSYSEQPRP
LMLPQDINELPYDKQLIFVQGTKTTPALKILARKIFYYEEEVFKSREKLPPPPLPVGDVSKIDALTVPVR
TIEAKVAVADAKPMQAEQRQRWNPRDTKTSATEVAQAEVDKAQPVEVEPDPEPVQADDTSEPM
MARVKSKPISPDNPQERIDEHPAFLLGKHPTEDSFLASYGQQFVMLAAPPGTGKGVGAVIPNLLSYPDSM
VVNDPKFENWEITSGFRASTGHKVYRFSPERLETHRWNPLSAINRDPLYRLGEIRTIARVLFVSDNPKNQ
EWYNKAGNVFTAMLLYLMETPEMPCTLPQAYEIGSLGTGIGTWAQQIIQLRSSGPNALSFETLRELNGVY
EASKNKSSGWSTTVDIVRDVLSVYAEKTVAWAVSGDDIDLSKAREEKMTAYFSVTEGSLKKYGPLMNLFF
TQAIRLNSKVIPEQGGHCADGTLRYKYQLALLMDELAIMGRIESLETAPALTRGAGLRFFFIFQGKDQLR
AIYGEEAANAIMKAIHNEIVFAPGDIKLAEEYSRRLGNTTVRVHNQSLNRQKHEVGARGQTDSYSEQPRP
LMLPQDINELPYDKQLIFVQGTKTTPALKILARKIFYYEEEVFKSREKLPPPPLPVGDVSKIDALTVPVR
TIEAKVAVADAKPMQAEQRQRWNPRDTKTSATEVAQAEVDKAQPVEVEPDPEPVQADDTSEPM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 48589..58981
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTL68_RS00260 | 43974..44501 | + | 528 | WP_032618636 | DUF2778 domain-containing protein | - |
| HTL68_RS00265 | 44501..44788 | + | 288 | WP_024670603 | hypothetical protein | - |
| HTL68_RS00270 | 44816..45589 | - | 774 | WP_024670604 | helix-turn-helix transcriptional regulator | - |
| HTL68_RS00275 | 45698..46798 | + | 1101 | WP_024670605 | polyamine ABC transporter substrate-binding protein | - |
| HTL68_RS00280 | 46895..47092 | + | 198 | WP_080268214 | hypothetical protein | - |
| HTL68_RS00285 | 47110..47496 | - | 387 | WP_172689638 | helix-turn-helix transcriptional regulator | - |
| HTL68_RS00290 | 47712..48245 | + | 534 | WP_122326351 | transcription termination/antitermination NusG family protein | - |
| HTL68_RS00295 | 48334..48576 | + | 243 | WP_172689639 | hypothetical protein | - |
| HTL68_RS00300 | 48589..49323 | + | 735 | WP_122326349 | lytic transglycosylase domain-containing protein | virB1 |
| HTL68_RS00305 | 49323..49658 | + | 336 | WP_024651578 | hypothetical protein | - |
| HTL68_RS00310 | 49671..50159 | + | 489 | WP_122326348 | VirB3 family type IV secretion system protein | virB3 |
| HTL68_RS00315 | 50062..52560 | + | 2499 | WP_235671738 | conjugal transfer protein | virb4 |
| HTL68_RS00320 | 52557..53243 | + | 687 | WP_024671760 | P-type DNA transfer protein VirB5 | - |
| HTL68_RS00325 | 53275..53628 | + | 354 | WP_024671759 | hypothetical protein | - |
| HTL68_RS00330 | 53639..54571 | + | 933 | WP_122326347 | type IV secretion system protein | virB6 |
| HTL68_RS00335 | 54981..55769 | + | 789 | WP_024671757 | type IV secretion system protein | virB8 |
| HTL68_RS00340 | 55759..56568 | + | 810 | WP_024671756 | P-type conjugative transfer protein VirB9 | virB9 |
| HTL68_RS00345 | 56555..57913 | + | 1359 | WP_122326346 | TrbI/VirB10 family protein | virB10 |
| HTL68_RS00350 | 57923..58981 | + | 1059 | WP_024668917 | P-type DNA transfer ATPase VirB11 | virB11 |
| HTL68_RS00355 | 58991..59347 | + | 357 | WP_024668916 | hypothetical protein | - |
| HTL68_RS00360 | 59473..59715 | + | 243 | WP_024668915 | hypothetical protein | - |
| HTL68_RS00365 | 59715..61376 | + | 1662 | WP_122326400 | type IV secretory system conjugative DNA transfer family protein | - |
| HTL68_RS00370 | 61416..61778 | + | 363 | WP_024668911 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HTL68_RS00375 | 61806..63971 | + | 2166 | WP_122335090 | type IA DNA topoisomerase | - |
Host bacterium
| ID | 3700 | GenBank | NZ_KY362367 |
| Plasmid name | pPg2708 | Incompatibility group | - |
| Plasmid size | 72672 bp | Coordinate of oriT [Strand] | 67391..67559 [+] |
| Host baterium | Pseudomonas coronafaciens pv. garcae strain 2708 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |