Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | ldrD-rdlD/Ldr(toxin) |
| Location | 4451656..4451878 | Replicon | chromosome |
| Accession | NZ_CP087294 | ||
| Organism | Escherichia coli strain APEC E956 | ||
Toxin (Protein)
| Gene name | ldrD | Uniprot ID | A0A829L523 |
| Locus tag | LO740_RS21625 | Protein ID | WP_000170738.1 |
| Coordinates | 4451656..4451763 (-) | Length | 36 a.a. |
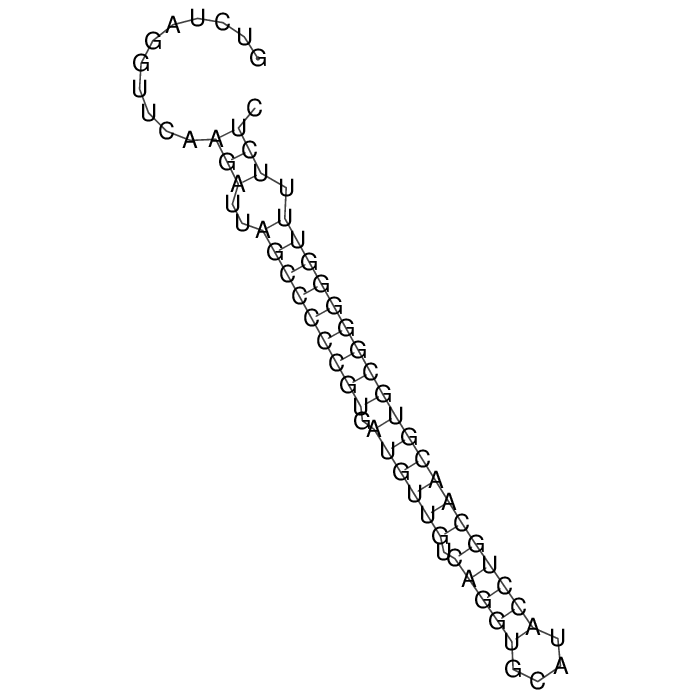
Antitoxin (RNA)
| Gene name | rdlD | ||
| Locus tag | - | ||
| Coordinates | 4451812..4451878 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LO740_RS21595 | 4446685..4448256 | + | 1572 | WP_001204944.1 | cellulose biosynthesis c-di-GMP-binding protein BcsE | - |
| LO740_RS21600 | 4448253..4448444 | + | 192 | WP_000988308.1 | cellulose biosynthesis protein BcsF | - |
| LO740_RS21605 | 4448441..4450120 | + | 1680 | WP_001562264.1 | cellulose biosynthesis protein BcsG | - |
| LO740_RS21610 | 4450207..4450314 | - | 108 | WP_001295224.1 | type I toxin-antitoxin system toxin Ldr family protein | - |
| LO740_RS21615 | 4450690..4450797 | - | 108 | WP_001295224.1 | type I toxin-antitoxin system toxin Ldr family protein | - |
| LO740_RS21620 | 4451173..4451280 | - | 108 | WP_001295224.1 | type I toxin-antitoxin system toxin Ldr family protein | - |
| LO740_RS21625 | 4451656..4451763 | - | 108 | WP_000170738.1 | type I toxin-antitoxin system toxin Ldr family protein | Toxin |
| - | 4451812..4451878 | + | 67 | - | - | Antitoxin |
| LO740_RS21630 | 4452239..4453510 | + | 1272 | WP_001318103.1 | aromatic amino acid transport family protein | - |
| LO740_RS21635 | 4453540..4454544 | - | 1005 | WP_000107036.1 | dipeptide ABC transporter ATP-binding subunit DppF | - |
| LO740_RS21640 | 4454541..4455524 | - | 984 | WP_001196486.1 | dipeptide ABC transporter ATP-binding protein | - |
| LO740_RS21645 | 4455535..4456437 | - | 903 | WP_000084677.1 | dipeptide ABC transporter permease DppC | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 36 a.a. Molecular weight: 3864.67 Da Isoelectric Point: 9.0157
>T225327 WP_000170738.1 NZ_CP087294:c4451763-4451656 [Escherichia coli]
MTLAELGMAFWHDLAAPVIAGILASLIVNWLNKRK
MTLAELGMAFWHDLAAPVIAGILASLIVNWLNKRK
Download Length: 108 bp
>T225327 NZ_CP121133:c3298172-3298070 [Klebsiella pneumoniae]
GCAAGGCGACTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
GCAAGGCGACTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
Antitoxin
Download Length: 67 bp
>AT225327 NZ_CP087294:4451812-4451878 [Escherichia coli]
GTCTAGGTTCAAGATTAGCCCCCGTGATGTTGTCAGGTGCATACCTGCAACGTGCGGGGGTTTTCTC
GTCTAGGTTCAAGATTAGCCCCCGTGATGTTGTCAGGTGCATACCTGCAACGTGCGGGGGTTTTCTC
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|