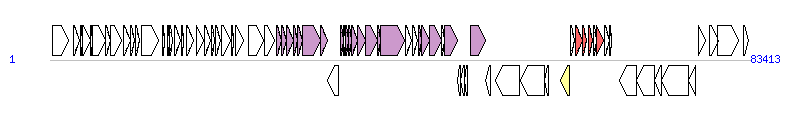The graph information of ICE6440 components from MF168944 |
 |
| Complete gene list of ICE6440 from MF168944 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 316..2244 [+], 1929 | Integrase | |
| 2 | - | 2796..3551 [+], 756 | Hypothetical protein | |
| 3 | - | 3670..3882 [+], 213 | Phage transcriptional regulator AlpA | |
| 4 | - | 3925..4800 [+], 876 | ParA | |
| 5 | - | 4784..5032 [+], 249 | Hypothetical protein | |
| 6 | - | 5025..6653 [+], 1629 | Putative ParB | |
| 7 | - | 6669..7229 [+], 561 | Hypothetical protein | |
| 8 | - | 7233..8477 [+], 1245 | Hypothetical protein | |
| 9 | - | 8795..9580 [+], 786 | Integrating conjugative element protein | |
| 10 | - | 9577..10104 [+], 528 | Hypothetical protein | |
| 11 | - | 10178..10618 [+], 441 | Single-stranded DNA-binding protein | |
| 12 | - | 10888..12912 [+], 2025 | DNA topoisomerase III | |
| 13 | - | 13437..13682 [+], 246 | Hypothetical protein | |
| 14 | - | 14011..14223 [+], 213 | Hypothetical protein | |
| 15 | - | 14245..14637 [+], 393 | Hypothetical protein | |
| 16 | - | 14850..15587 [+], 738 | Hypothetical protein | |
| 17 | - | 15682..15960 [+], 279 | Hypothetical protein | |
| 18 | - | 16240..17052 [+], 813 | Hypothetical protein | |
| 19 | - | 17430..18344 [+], 915 | Hypothetical protein | |
| 20 | - | 18399..19097 [+], 699 | Hypothetical protein | |
| 21 | - | 19192..19593 [+], 402 | Hypothetical protein | |
| 22 | - | 19676..20326 [+], 651 | Hypothetical protein | |
| 23 | - | 20391..21512 [+], 1122 | Hypothetical protein | |
| 24 | - | 21613..21924 [+], 312 | Hypothetical protein | |
| 25 | - | 22062..23117 [+], 1056 | DEAD/DEAH box helicase | |
| 26 | - | 23713..25431 [+], 1719 | Group II intron reverse transcriptase/maturase | |
| 27 | - | 25598..26758 [+], 1161 | DEAD/DEAH box helicase | |
| 28 | - | 26992..27570 [+], 579 | Conj_PilL superfamily protein | Tfc2, T4SS component |
| 29 | - | 27567..28208 [+], 642 | Hypothetical protein | Tfc2, T4SS component |
| 30 | - | 28223..28960 [+], 738 | Integrating conjugative element protein | Tfc3, T4SS component |
| 31 | - | 28960..29547 [+], 588 | Lytic transglycosylase | Tfc4, T4SS component |
| 32 | - | 29544..30092 [+], 549 | Integrating conjugative element protein | Tfc5, T4SS component |
| 33 | - | 30097..32283 [+], 2187 | Conjugative coupling factor TraD, PFGI-1 class | Tfc6, T4SS component |
| 34 | - | 32280..33029 [+], 750 | Integrating conjugative element membrane protein | Tfc8, T4SS component |
| 35 | - | 33030..34400 [-], 1371 | hypothetical protein | |
| 36 | - | 34562..34942 [+], 381 | Integrative conjugative element protein | Tfc9, T4SS component |
| 37 | - | 34939..35172 [+], 234 | integrating conjugative element protein | |
| 38 | - | 35189..35548 [+], 360 | integrating conjugative element membrane protein | Tfc10, T4SS component |
| 39 | - | 35561..35971 [+], 411 | Conjugative transfer region protein | Tfc11, T4SS component |
| 40 | - | 35968..36657 [+], 690 | integrating conjugative element protein | Tfc12, T4SS component |
| 41 | - | 36654..37586 [+], 933 | Integrating conjugative element protein | Tfc13, T4SS component |
| 42 | - | 37576..38994 [+], 1419 | Integrating conjugative element protein | Tfc14, T4SS component |
| 43 | - | 38975..39415 [+], 441 | Conjugal transfer protein | Tfc15, T4SS component |
| 44 | - | 39415..42285 [+], 2871 | Conjugative transfer ATPase | Tfc16, T4SS component |
| 45 | - | 42328..43053 [+], 726 | DSBA oxidoreductase | |
| 46 | - | 43240..43734 [+], 495 | DNA repair protein RadC | |
| 47 | - | 43878..44324 [+], 447 | Integrating conjugative element protein | Tfc24, T4SS component |
| 48 | - | 44321..45268 [+], 948 | integrating conjugative element protein | Tfc23, T4SS component |
| 49 | - | 45279..46673 [+], 1395 | integrating conjugative element protein | Tfc22, T4SS component |
| 50 | - | 46670..47029 [+], 360 | Hypothetical protein | |
| 51 | - | 47045..48568 [+], 1524 | conjugal transfer protein TraG | Tfc19, T4SS component |
| 52 | - | 48575..48937 [-], 363 | Hypothetical protein | |
| 53 | - | 48965..49423 [-], 459 | toxin YhaV | |
| 54 | - | 49423..49740 [-], 318 | AbrB family transcriptional regulator | |
| 55 | - | 50052..51914 [+], 1863 | Integrating conjugative element relaxase | Relaxase, MOBH Family |
| 56 | - | 51925..52506 [-], 582 | Hypothetical protein | |
| 57 | - | 53075..55960 [-], 2886 | Transposase | |
| 58 | - | 56109..58868 [-], 2760 | Transposase | |
| 59 | - | 58871..59431 [-], 561 | TnpR resolvase | |
| 60 | - | 60844..61857 [-], 1014 | IntI1 integrase of class I integrons | Integrase |
| 61 | - | 62004..62462 [+], 459 | Aminoglycoside N-acetyltransferase AAC(6')-Il | |
| 62 | - | 62615..63415 [+], 801 | VIM-2 carbapenemase | AR |
| 63 | - | 63509..63973 [+], 465 | AAC(3)-I family aminoglycoside 3-N-acetyltransferase | AR |
| 64 | - | 64130..64648 [+], 519 | AAC(6')-Ib family aminoglycoside 6'-N-acetyltransferase | AR |
| 65 | - | 64817..65164 [+], 348 | QacEdelta1 multidrug efflux protein | |
| 66 | - | 65158..65997 [+], 840 | Sul1 sulfonamide-resistant dihydropteroate synthase | AR |
| 67 | - | 66029..66625 [+], 597 | Orf5 acetyltransferase | |
| 68 | - | 66649..66936 [+], 288 | Orf6 protein | |
| 69 | - | 67868..69889 [-], 2022 | site-specific DNA-methyltransferase | |
| 70 | - | 69889..72012 [-], 2124 | hypothetical protein | |
| 71 | - | 72022..72828 [-], 807 | hypothetical protein | |
| 72 | - | 72825..76046 [-], 3222 | helicase | |
| 73 | - | 76059..76967 [-], 909 | WYL domain-containing protein | |
| 74 | - | 77247..78137 [+], 891 | LysR family transcriptional regulator | |
| 75 | - | 78582..79613 [+], 1032 | ATP-binding protein | |
| 76 | - | 79591..82101 [+], 2511 | Putative serine protease | |
| 77 | - | 82627..83268 [+], 642 | LysR family transcriptional regulator | |