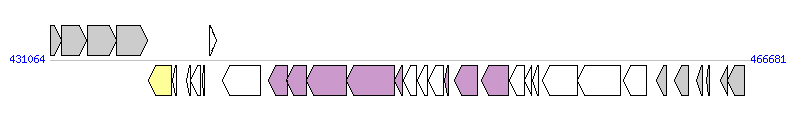The graph information of Tn5801 components from BA000017 |
 |
| Complete gene list of Tn5801 from BA000017 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | xprT | 431087..431665 [+], 579 | xanthine phosphoribosyltransferase | |
| 2 | pbuX | 431665..432933 [+], 1269 | xanthine permease | |
| 3 | guaB | 432971..434437 [+], 1467 | inositol-monophosphate dehydrogenase | |
| 4 | guaA | 434462..436003 [+], 1542 | GMP synthase | |
| 5 | int | 436071..437264 [-], 1194 | integrase homolog | Integrase |
| 6 | SAV0393 | 437291..437491 [-], 201 | hypothetical protein | |
| 7 | SAV0394 | 437989..438219 [-], 231 | conserved hypothetical protein | |
| 8 | SAV0395 | 438216..438698 [-], 483 | hypothetical protein | |
| 9 | SAV0396 | 438827..438937 [-], 111 | hypothetical protein | |
| 10 | SAV0397 | 439169..439522 [+], 354 | putative transcriptional regulator | |
| 11 | tetM | 439865..441784 [-], 1920 | tetracycline resistance protein | |
| 12 | SAV0399 | 442161..443123 [-], 963 | hypothetical protein | Orf13_Tn, T4SS component |
| 13 | SAV0400 | 443107..444129 [-], 1023 | similar to lipoprotein, NLP/P60 family | Orf14_Tn, T4SS component |
| 14 | SAV0401 | 444126..446153 [-], 2028 | putative membrane protein | Orf15_Tn, T4SS component |
| 15 | SAV0402 | 446150..448603 [-], 2454 | hypothetical protein | Orf16_Tn, T4SS component |
| 16 | SAV0403 | 448587..449024 [-], 438 | hypothetical protein | Orf17_Tn, T4SS component |
| 17 | SAV0404 | 449054..449692 [-], 639 | hypothetical protein | |
| 18 | SAV0405 | 449748..450248 [-], 501 | hypothetical protein | |
| 19 | SAV0406 | 450309..451088 [-], 780 | hypothetical protein | |
| 20 | SAV0407 | 451130..451351 [-], 222 | hypothetical protein | Orf19_Tn, T4SS component |
| 21 | SAV0408 | 451634..452818 [-], 1185 | putative phage replication protein | |
| 22 | SAV0409 | 453000..454403 [-], 1404 | similar to DNA translocase FtsK | Orf21_Tn, T4SS component |
| 23 | SAV0410 | 454425..455198 [-], 774 | hypothetical protein | |
| 24 | SAV0411 | 455208..455585 [-], 378 | hypothetical protein | |
| 25 | SAV0412 | 455606..455920 [-], 315 | hypothetical protein | |
| 26 | SAV0413 | 456120..457913 [-], 1794 | hypothetical protein | |
| 27 | SAV0414 | 457910..460099 [-], 2190 | hypothetical protein | |
| 28 | tnp | 460233..461423 [-], 1191 | transposase | |
| 29 | SAV0416 | 461917..462453 [-], 537 | hypothetical protein | |
| 30 | SAV0417 | 462845..463549 [-], 705 | hypothetical protein | |
| 31 | SAV0418 | 463948..464244 [-], 297 | probable transposase | |
| 32 | SAV0419 | 464481..464633 [-], 153 | hypothetical protein | |
| 33 | SAV0420 | 465165..465524 [-], 360 | conserved hypothetical protein | |
| 34 | SAV0421 | 465543..466388 [-], 846 | putative nucleoside-diphosphate-sugar epimerase | |